Bottom-Up MS-Based Post-Translational Modification Analysis Service
MtoZ Biolabs provides Bottom-Up MS-Based Post-Translational Modification (PTM) Analysis Service to support large-scale, sensitive, and quantitative analysis of protein modifications across diverse biological samples. The bottom-up approach remains the most widely adopted strategy in proteomics due to its robustness, sensitivity, and compatibility with high-throughput workflows.
By integrating optimized sample preparation, targeted enrichment, advanced LC-MS/MS instrumentation, and intelligent bioinformatics, MtoZ Biolabs delivers reliable datasets that characterize site-specific PTMs with exceptional coverage and reproducibility. Our service enables researchers to explore complex molecular networks and uncover the biological significance of PTM regulation in both physiological and pathological contexts.
Technical Principles
The bottom-up mass spectrometry approach is a peptide-centered analytical strategy that identifies and quantifies proteins through their digested peptide fragments. Proteins are enzymatically cleaved into shorter peptides, which are then separated, ionized, and analyzed by tandem mass spectrometry.
Each peptide produces a distinct fragmentation spectrum that enables accurate sequence determination and precise localization of modification sites. Enzymatic digestion, most commonly performed using trypsin or Lys-C, provides consistent and comprehensive peptide coverage. To enhance the detection of low-abundance PTMs, enrichment procedures such as IMAC, TiO₂, HILIC, or antibody affinity are employed before LC-MS/MS analysis.
Quantitative strategies including TMT, iTRAQ, and label-free methods allow accurate comparison of PTM abundance across biological samples or treatment groups. These combined analytical principles provide a high-resolution view of protein modification dynamics and reveal the functional relevance of PTM-mediated regulation.
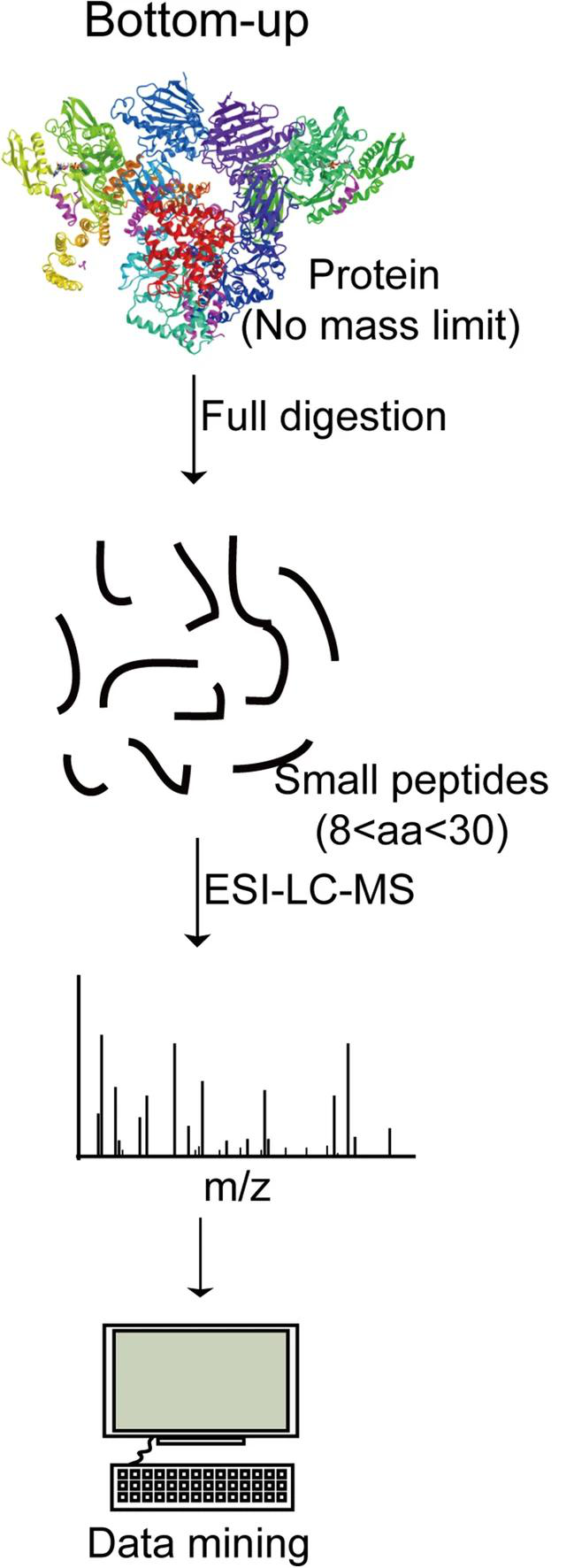
Sun, M. et al. Cell Biosci. 2022.
Figure 1. Schematic Workflow of Bottom-Up Proteomics
Analysis Workflow
Bottom-Up MS-Based Post-Translational Modification Analysis Service at MtoZ Biolabs integrates these principles into a unified workflow that maximizes data accuracy, reproducibility, and biological insight.
1. Sample Preparation: Extraction of proteins under optimized conditions to preserve modification integrity.
2. Enzymatic Digestion: Proteins are cleaved into peptides using high-specificity proteases.
3. Targeted Peptide Enrichment: Specific enrichment strategies (IMAC, TiO₂, HILIC, SCX, or antibody affinity) are applied to isolate PTM-containing peptides.
4. LC-MS/MS Analysis: High-resolution Orbitrap or timsTOF MS systems acquire fragmentation spectra for confident PTM site identification.
5. Quantitative Profiling: TMT, iTRAQ, or label-free methods enable comparative analysis across experimental conditions.
6. Bioinformatics Interpretation: Data are processed through advanced software pipelines for PTM mapping, functional annotation, and pathway analysis.
Why Choose MtoZ Biolabs?
✅ Extensive PTM Expertise: Decades of experience in proteomics and post-translational modification analysis.
✅ High Sensitivity and Reproducibility: State-of-the-art LC-MS/MS platforms ensure deep proteome coverage and accurate quantification.
✅ Specialized Enrichment Techniques: Optimized protocols for detecting low-abundance and labile PTMs.
✅ Integrated Bioinformatics: Complete analytical pipeline from acquisition to biological interpretation.
✅ Customized Analytical Design: Flexible project planning and expert consultation tailored to individual research goals.
✅ One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
Sample Submission Suggestions
MtoZ Biolabs accepts a wide range of biological materials suitable for bottom-up MS analysis:
Cells: Minimum of 1 × 10⁷ cells.
Animal or Human Tissue: ≥200 mg, stored at -80°C.
Serum or Plasma: ≥100 µL.
Urine or Culture Supernatant: ≥2 mL.
Purified Proteins: ≥20 µg, detergent- and salt-free.
Samples should be shipped on dry ice. For complex matrices or specialized enrichment requirements, our technical experts provide pre-analysis consultation to ensure optimal outcomes.
Applications
The Bottom-Up MS-Based Post-Translational Modification Analysis Service supports a wide range of biological and translational applications:
Cell Signaling and Pathway Analysis: Identification of phosphorylation and ubiquitination events involved in signaling regulation.
Epigenetic and Chromatin Studies: Comprehensive mapping of histone PTMs influencing gene expression and chromatin remodeling.
Disease Mechanism Research: Detection of PTM alterations associated with cancer, neurodegenerative, and metabolic disorders.
Drug Mechanism Evaluation: Assessing PTM dynamics in response to therapeutic compound treatment.
Biomarker Discovery: Identifying PTM-based molecular markers for disease diagnosis and treatment monitoring.
Bottom-Up MS-Based Post-Translational Modification Analysis Service at MtoZ Biolabs provides a robust and scalable platform for high-resolution PTM characterization. Our optimized workflows, advanced instrumentation, and integrated bioinformatics ensure precise identification and quantitative interpretation of protein modifications.
To learn how MtoZ Biolabs can support your PTM research or to request a customized experimental plan, please contact our scientific team for consultation.







