Histone Monomethylation Analysis Service
Based on a high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS) platform combined with specific modified peptide enrichment methods, the histone monomethylation analysis service launched by MtoZ Biolabs enables precise detection and quantitative analysis of monomethylation modifications on lysine or arginine residues of histones. This service can identify monomethylation modifications at different sites and monitor their abundance changes under different experimental conditions. By integrating bioinformatics approaches, it provides high-quality data on modification site distribution, differential comparison, and functional annotation, offering reliable support for in-depth analysis of the functional characteristics of monomethylation.
Overview
Histone monomethylation (me1) refers to a type of post-translational modification in which a single methyl group is attached to lysine or arginine residues of histones, mainly occurring on the N-terminal tails of histones H3 and H4, and it plays an important role in maintaining chromatin structure and regulating gene transcription. Monomethylation is critical in biological processes such as chromatin remodeling, gene expression regulation, cell cycle control, as well as development and differentiation. Histone monomethylation analysis enables systematic detection and quantitative evaluation of monomethylation modifications at different sites, with broad applications in epigenetic research, cellular function studies, and potential biomarker discovery, providing reliable data support for understanding the role of monomethylation in life processes.
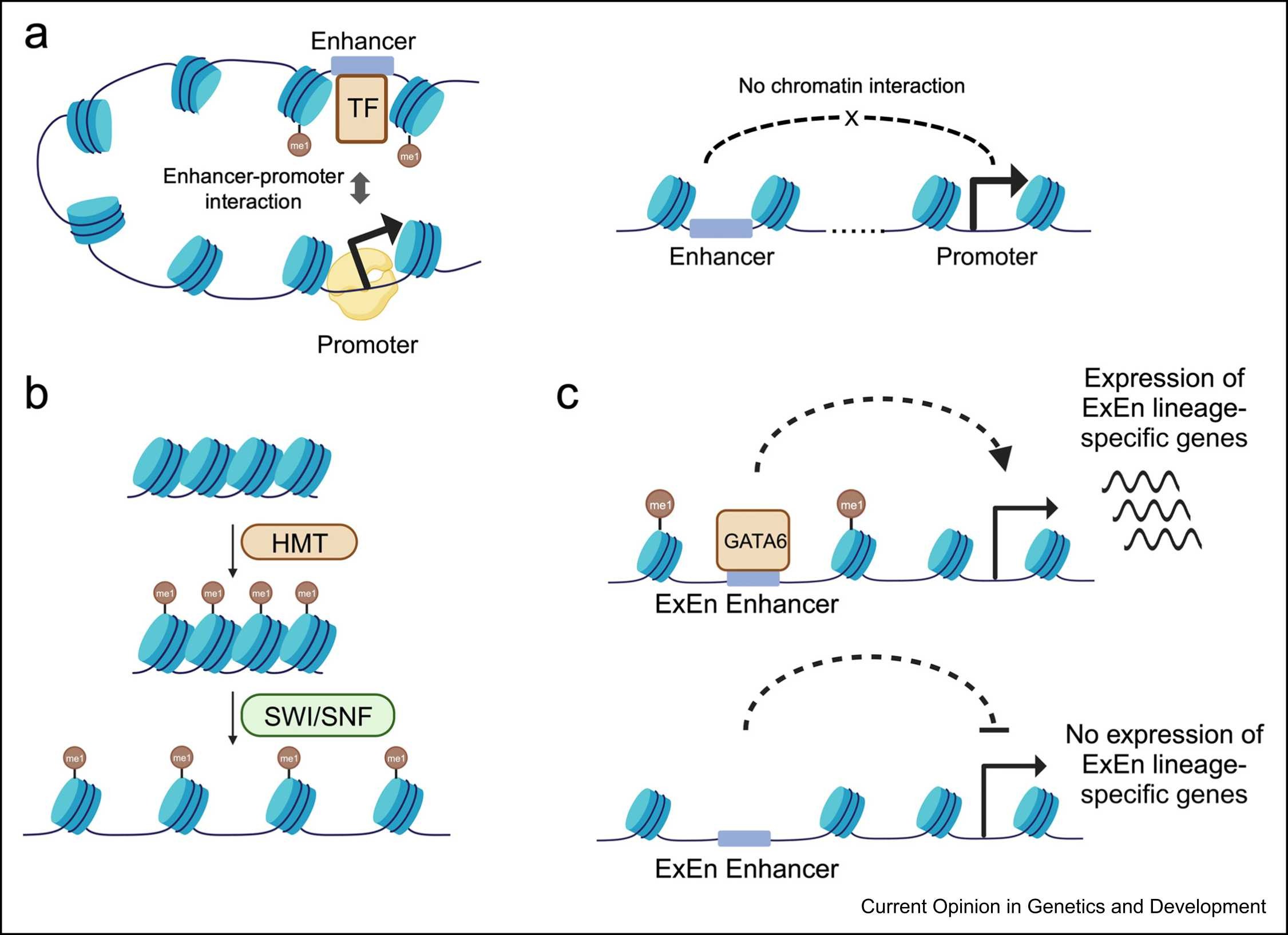
Wang, Z N. et al. Current Opinion in Genetics & Development, 2024.
Figure 1. Models on the Roles of H3K4me1 in Gene Regulation.
Analysis Workflow
1. Histone Extraction and Digestion
Histones are isolated from cell or tissue samples and digested under optimized conditions to generate detectable peptides.
2. Modified Peptide Enrichment
Specific enrichment methods are applied to capture monomethylated peptides, improving detection rate and coverage.
3. LC-MS/MS Analysis
Using a high-resolution liquid chromatography–tandem mass spectrometry platform, monomethylation sites are accurately identified and quantified.
4. Data Analysis
Bioinformatics tools are integrated to provide modification distribution and abundance differences, supporting functional studies.
Sample Submission Suggestions
1. Sample Type and Quantity
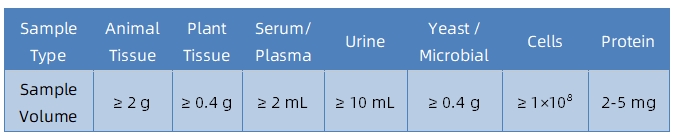
Note: Plasma should be collected using EDTA as an anticoagulant. Standard tissue or cell lysis buffers can be used during protein extraction.
2. Sample Transportation
Avoid repeated freeze-thaw cycles. Samples are recommended to be stored at -80°C and transported on dry ice to ensure low-temperature conditions throughout the process and prevent modification loss.
Note: For special samples or if a detailed submission plan is required, please contact MtoZ Biolabs technical staff in advance.
Service Advantages
1. High-Resolution Detection
Relying on an advanced LC-MS/MS platform to ensure precise identification of low-abundance monomethylation modifications.
2. Strict Quality Control
Standardized workflow with multiple quality control steps guarantees stability and reproducibility of results.
3. One-Stop Service Workflow
An integrated solution covering sample preparation, mass spectrometry detection, and data analysis.
4. Flexible Workflow Design
Experimental procedures are customized based on sample type and research objectives to meet diverse needs.
Applications
1. Epigenetics Research
Histone monomethylation analysis service can be used to analyze the role of monomethylation in chromatin structural changes and gene transcription regulation.
2. Cell Differentiation and Development Research
By studying monomethylation modifications during stem cell differentiation and tissue development, its dynamic regulation can be explored.
3. Chromatin Remodeling Research
Histone monomethylation analysis service can be applied to investigate how monomethylation affects nucleosome stability and chromatin accessibility.
4. Potential Biomarker Discovery
By identifying monomethylation patterns under specific conditions, the service contributes to the discovery of novel biomarkers.
FAQ
Q1: Which Sites Can Histone Monomethylation Analysis Detect?
A1: This service can detect monomethylation sites on multiple histone subtypes (such as H3 and H4), including common ones like H3K4me1 and H3K9me1, and the detection range can be expanded based on research needs.
Q2: Can It Be Combined with Other Epigenetic Modification Analyses?
A2: Yes. Monomethylation often cooperates or antagonizes with modifications such as acetylation, dimethylation, or trimethylation. Combined analysis helps to reveal the interactions among different modifications.
Q3: Does the Service Support Customized Requirements?
A3: Yes. The experimental workflow and data analysis strategy can be flexibly adjusted according to research objectives and sample characteristics to meet personalized research needs.







