DIA-Based Glycoproteomics Service
At MtoZ Biolabs, the DIA-Based Glycoproteomics Service is designed to overcome the intrinsic complexity and heterogeneity of protein glycosylation by leveraging advanced data-independent acquisition strategies. Our platform integrates optimized glycopeptide enrichment, high-resolution LC-MS/MS on Orbitrap and Q-TOF systems, and tailored bioinformatics pipelines to ensure comprehensive identification and quantification of both N- and O-linked glycopeptides. By standardizing workflows while allowing project-specific customization, we provide clients with reproducible datasets and biologically meaningful insights that are directly applicable to disease mechanism studies, biomarker discovery, and therapeutic protein development.
Overview
Data-independent acquisition (DIA) has rapidly emerged as a transformative strategy in bottom-up proteomics and is increasingly applied to glycoproteomics, where traditional data-dependent acquisition (DDA) faces inherent limitations. DDA often underrepresents glycopeptides because they are typically of low abundance and ionization efficiency, resulting in poor coverage and inconsistent quantification. In contrast, DIA systematically fragments all precursor ions within predefined mass-to-charge windows, generating comprehensive and unbiased spectral libraries that capture signals from both abundant and rare glycopeptides.
The adoption of DIA in glycoproteomics significantly enhances sensitivity, reproducibility, and quantitative accuracy. It enables large-scale detection of glycoforms, reliable estimation of site occupancy, and improved assessment of glycosylation heterogeneity, even in complex biological matrices. These advantages make DIA particularly valuable for projects requiring deep coverage and precise quantification, including biomarker discovery, therapeutic protein characterization, and mechanistic studies of disease-associated glycosylation.
Technical Principles
DIA-Based Glycoproteomics relies on systematically fragmenting all precursor ions within predefined mass-to-charge windows during LC-MS/MS acquisition, ensuring comprehensive and unbiased coverage of glycopeptides. Each cycle generates multiplexed spectra that contain fragment information from both peptide backbones and attached glycans. These data-rich spectra are then processed using specialized algorithms and curated glycan databases to accurately assign peptide sequences, localize glycosylation sites, and annotate glycan structures. The approach enables simultaneous site-specific mapping and quantitative profiling of glycoform distributions, providing a detailed and reproducible characterization of protein glycosylation.
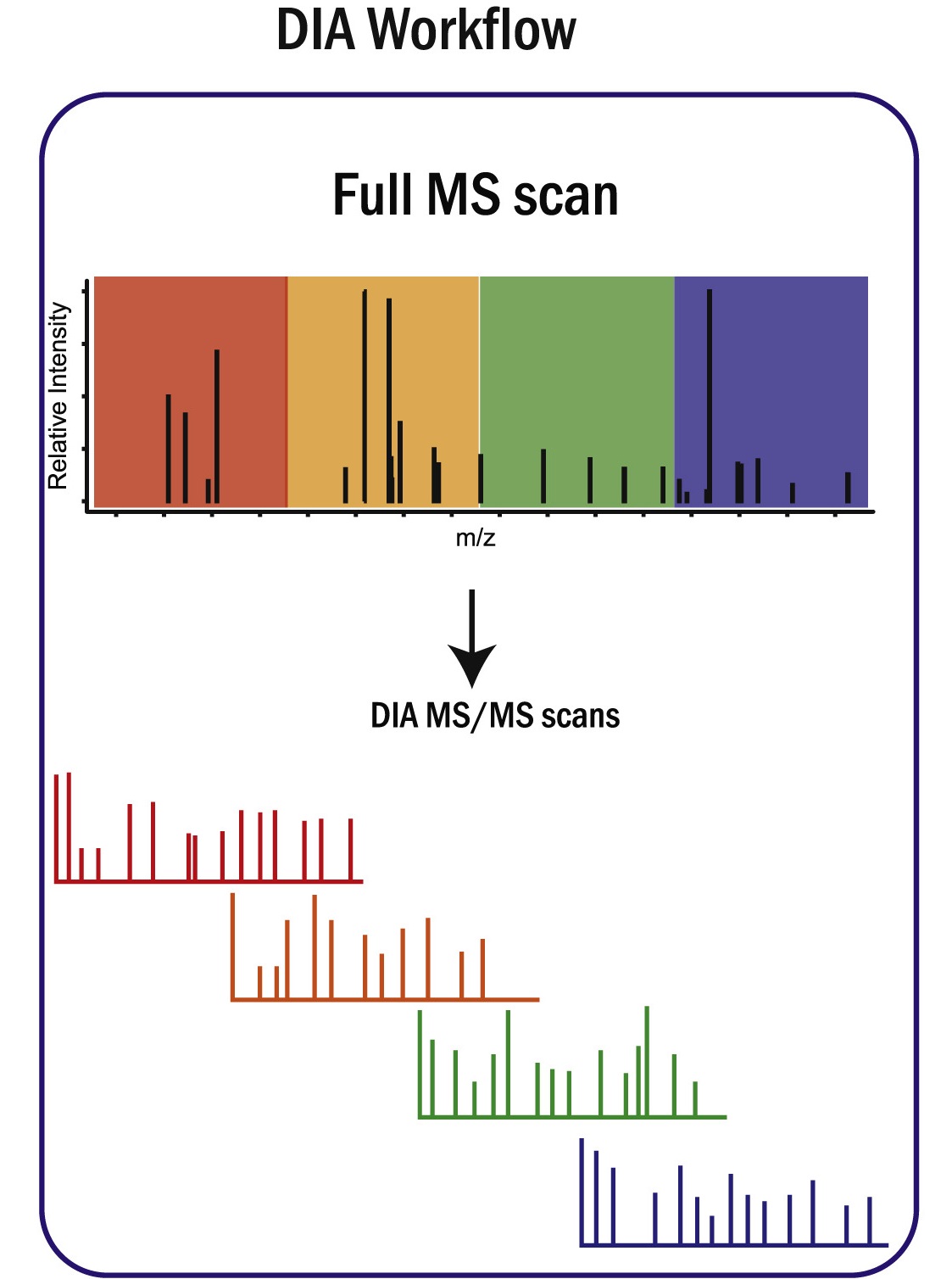
Ye Z, Vakhrushev SY. Mol Cell Proteomics. 2021.
Figure 1. Schematic Depiction of DIA Workflow
Analysis Workflow
The general analytical workflow for the DIA-Based Glycoproteomics Service is as follows:
1. Sample Preparation
Proteins are extracted and digested under optimized conditions to preserve glycosylation, followed by optional glycopeptide enrichment to improve coverage.
2. LC Separation
Peptides and glycopeptides are separated using high-performance liquid chromatography to reduce sample complexity and improve ionization efficiency.
3. DIA-MS Acquisition
All precursor ions within defined m/z windows are systematically fragmented in a DIA mode, generating comprehensive spectra containing both peptide and glycan fragment information.
4. Data Processing
Raw spectra are deconvoluted using specialized bioinformatics pipelines to assign peptide sequences, localize glycosylation sites, and annotate glycan structures.
5. Quantitative and Functional Analysis
Relative quantification of glycoform distributions and site occupancy is performed, followed by functional annotation and pathway mapping when required.
6. Reporting
Results are compiled into structured reports including glycopeptide identification, glycosylation site mapping, quantitative data, and annotated spectra.
Service Advantages
1. Deep and Comprehensive Coverage
Captures site-specific glycosylation and diverse glycoforms across complex biological samples.
2. Accurate Quantification
Provides reliable estimation of glycosylation site occupancy and precise glycoform distribution profiling.
3. High Reproducibility
Delivers consistent and reproducible datasets, ensuring robust results for both research and biopharmaceutical applications.
Sample Submission Suggestions
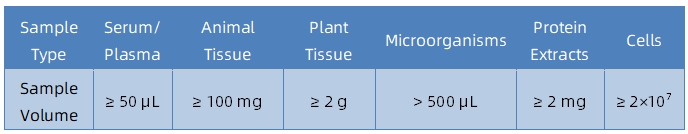
It is recommended to contact our technical support team before submitting samples to determine sample suitability and obtain tailored submission guidelines.
Applications
Applications of the DIA-Based Glycoproteomics Service include but are not limited to:
1. Therapeutic Protein Characterization
Site-specific analysis of glycosylation in monoclonal antibodies, fusion proteins, and other biologics to support quality control and regulatory compliance.
2. Biosimilarity Assessment
Comparative profiling of glycosylation patterns between biosimilar candidates and reference products to demonstrate structural and functional equivalence.
3. Biomarker Discovery
Identification of disease-associated glycosylation changes in serum, plasma, or tissue samples for early diagnosis and precision medicine.
4. Disease Mechanism Research
Investigation of glycosylation alterations in cancer, autoimmune disorders, neurodegeneration, and infectious diseases to reveal underlying molecular mechanisms.







