Protein Post-Translational Modifications Profiling Service
Based on advanced instrumentation and analytical platforms, MtoZ Biolabs has launched the protein post-translational modifications profiling service which enables systematic detection and characterization of multiple post-translational modifications (PTMs) in protein samples. This service integrates multidimensional separation technologies with high-sensitivity mass spectrometry acquisition to achieve comprehensive identification and quantification of diverse modifications. Through standardized data processing and bioinformatics analysis, it provides information on modification site distribution, abundance variations, and pathway associations, offering comprehensive and reliable data support for studying protein modification networks and dynamic regulatory mechanisms.
What Is Post-Translational Modification?
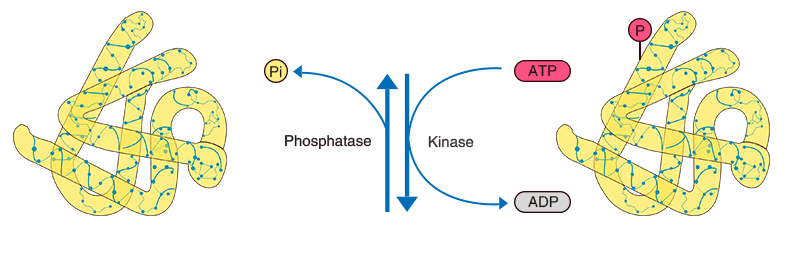
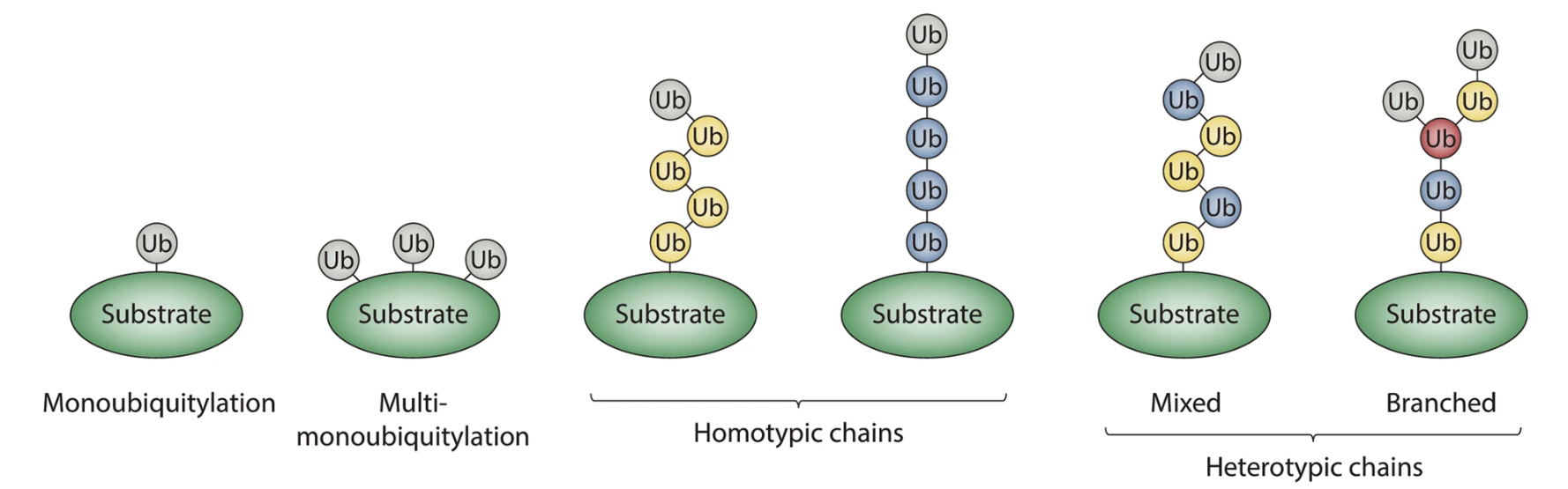
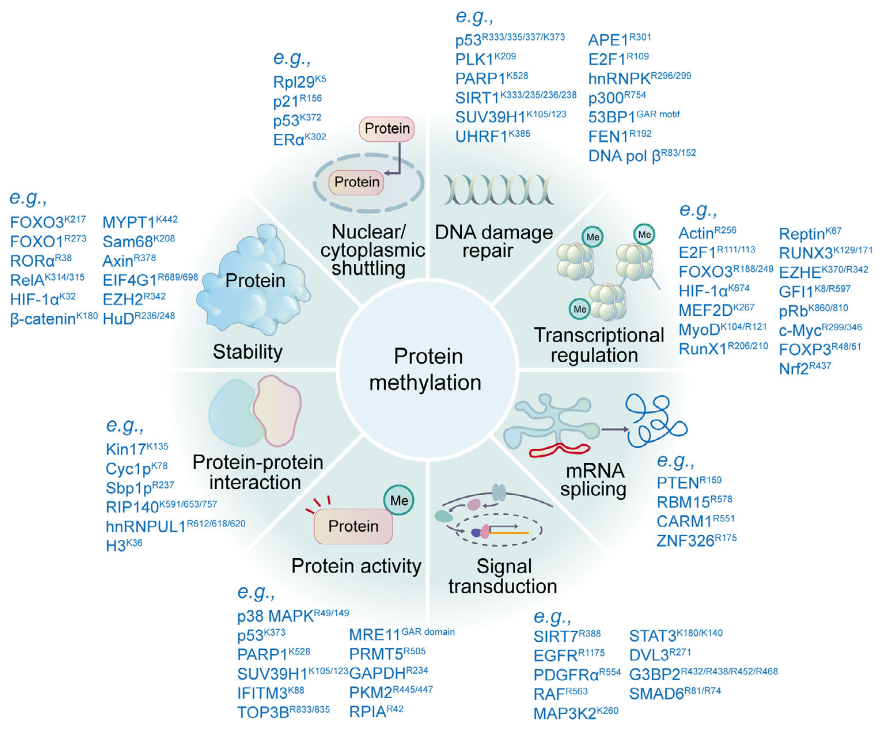
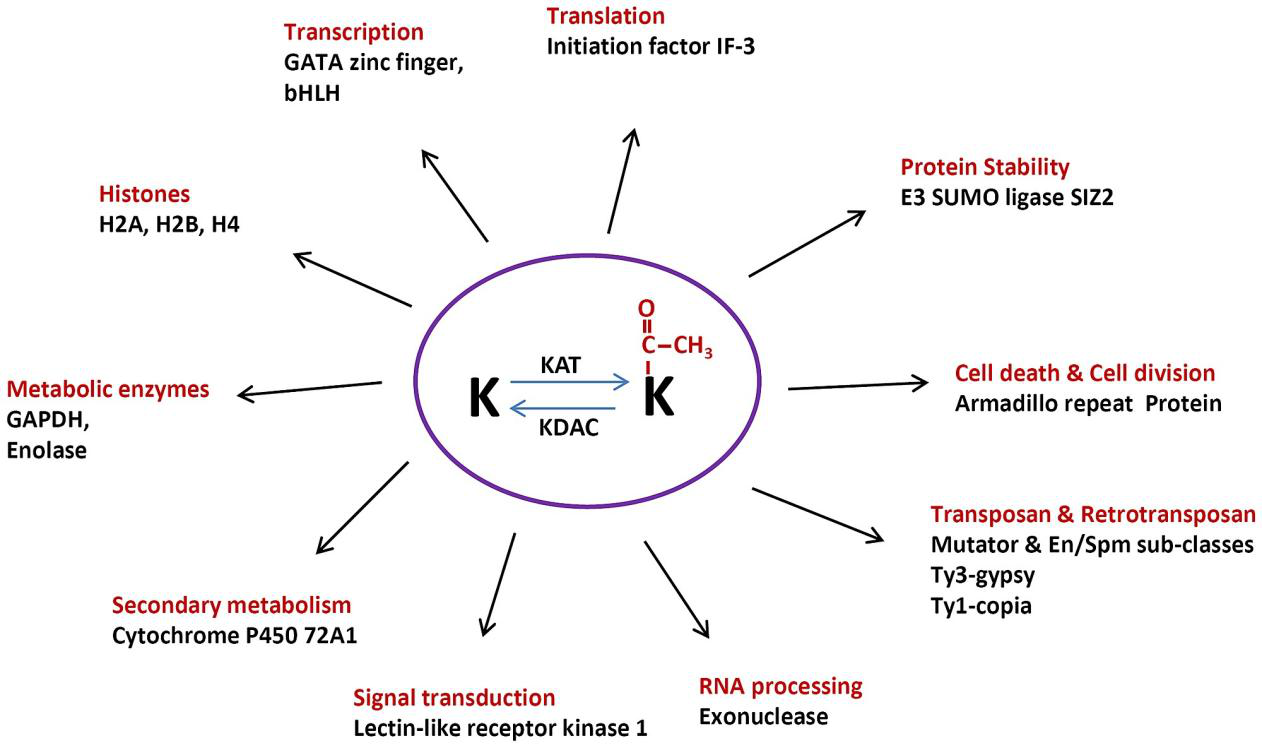
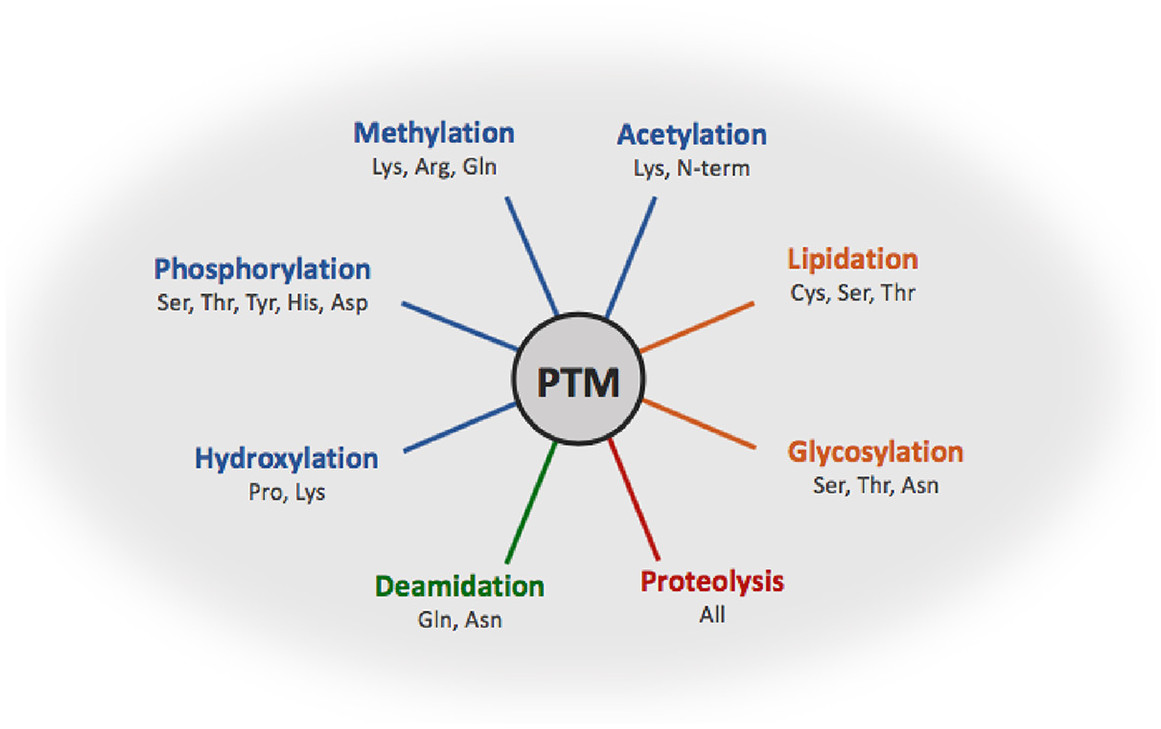
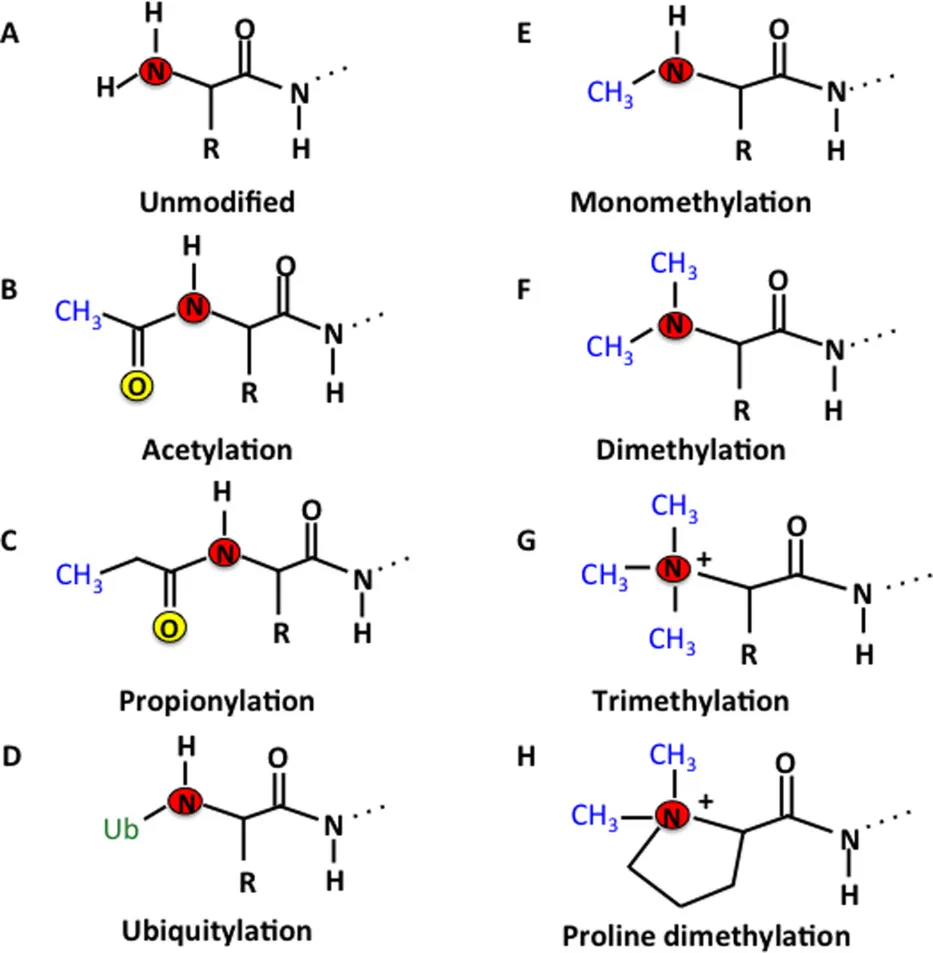
Protein post-translational modifications (PTMs) refer to structural alterations that occur on specific amino acid residues of proteins after translation through chemical or enzymatic reactions, such as phosphorylation, acetylation, methylation, ubiquitination, and glycosylation. These modifications regulate protein activity, conformation, interactions, and localization, thereby participating in cellular signaling and functional regulation. Accurate characterization of protein post-translational modifications is essential for understanding cellular functional networks and their dynamic changes. Protein post-translational modifications profiling can comprehensively reveal modification distribution and abundance variations under different conditions, providing key data support for studying cellular regulatory mechanisms, protein functional characteristics, and biological processes at the molecular level.
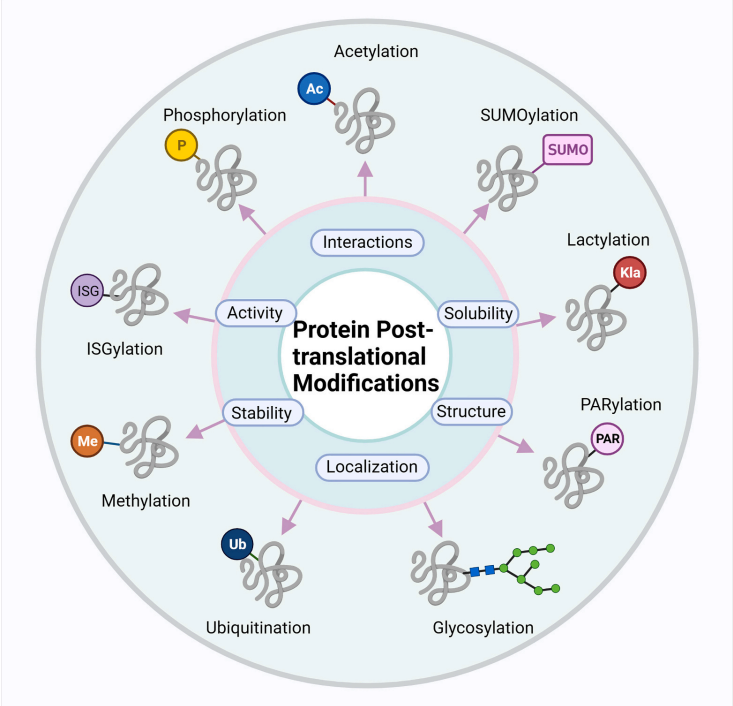
Chen, Y M. et al. International Journal of Biological Macromolecules, 2023.
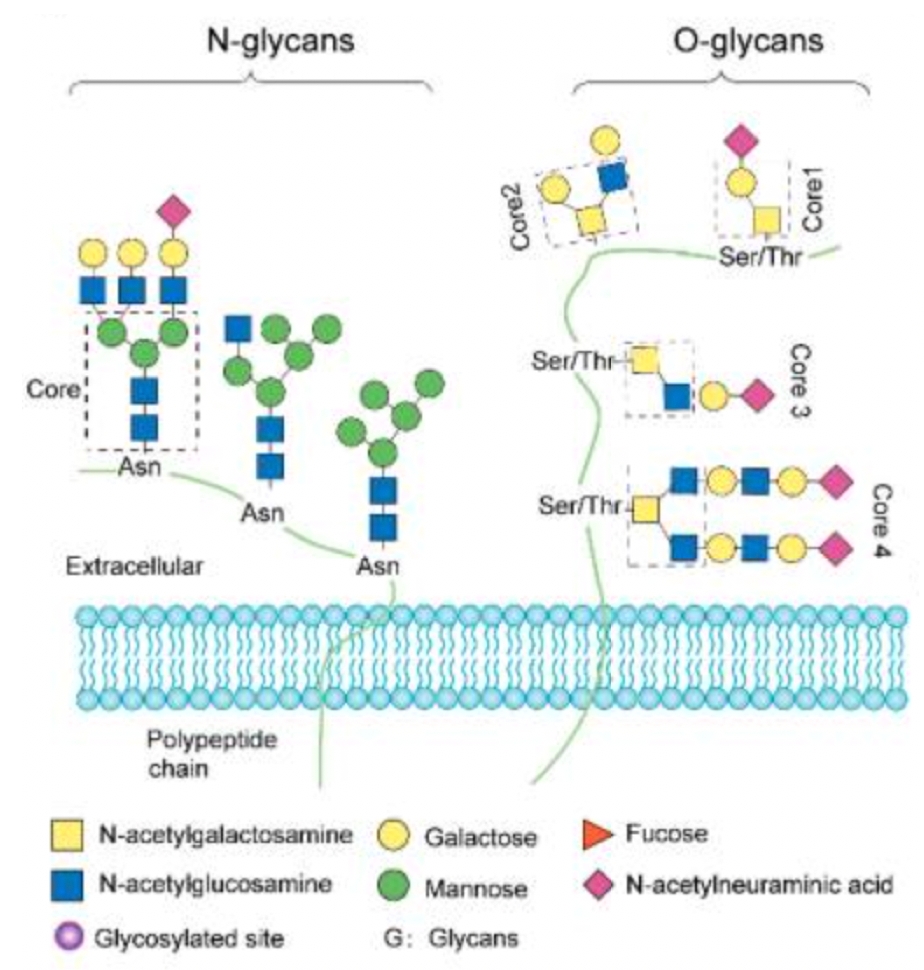
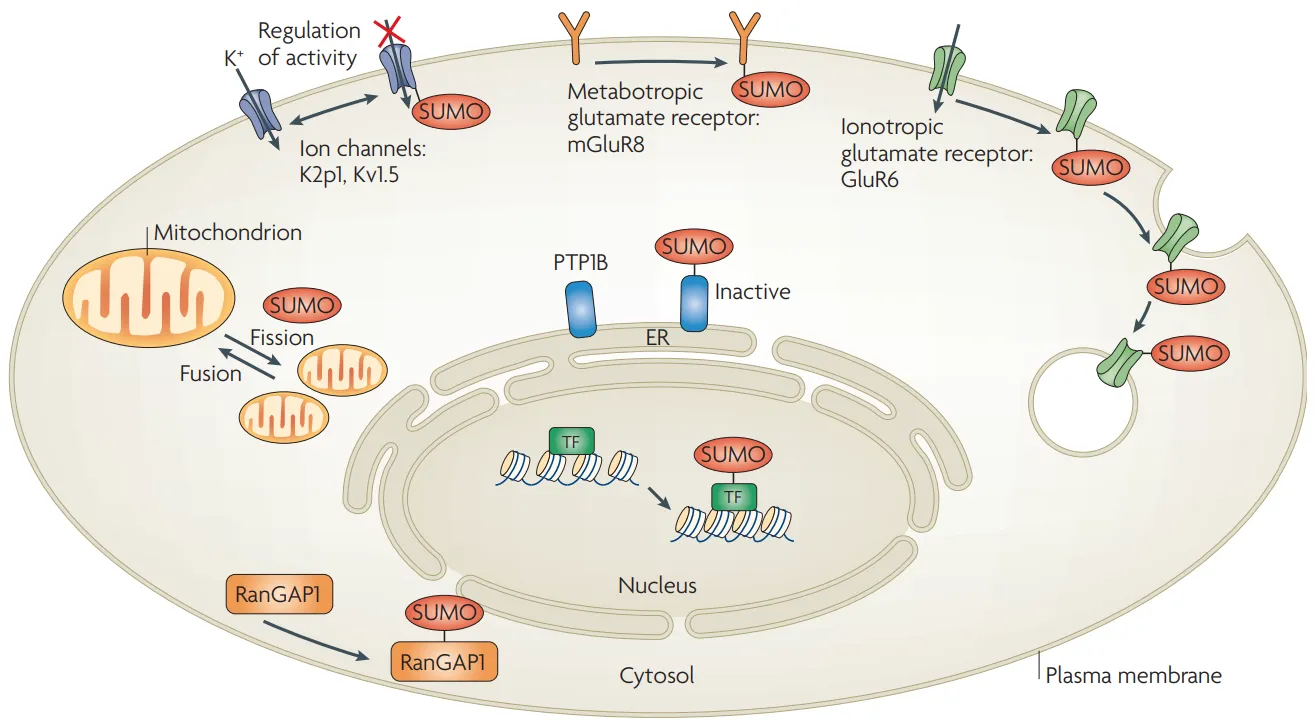
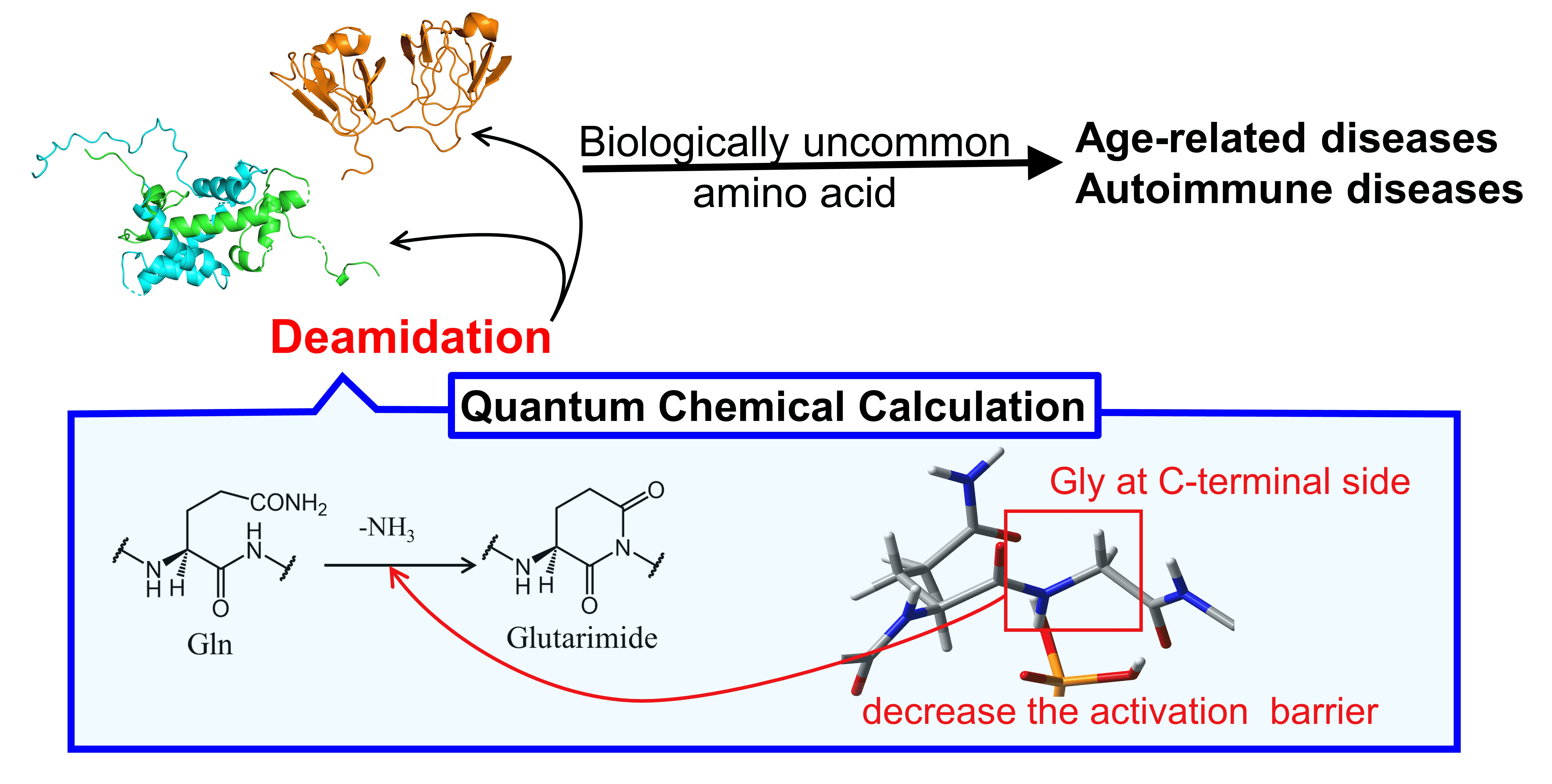
Figure. 1. The Fate of Proteins after Post-Translational Modifications (PTMs).
Services at MtoZ Biolabs
1. Target Proteins Post-Translational Modification Analysis
MtoZ Biolabs can perform post-translational modification analysis on specific target proteins, identifying modification types and sites. By combining high-resolution LC-MS/MS, we can compare modification changes under different conditions, providing data support for protein function regulation.
2. Proteomics Post-Translational Modification Analysis
By combining enrichment strategies with high-throughput mass spectrometry, MtoZ Biolabs can systematically analyze the post-translational modification information of numerous proteins at the proteomic level. This analysis helps reveal global PTM patterns and supports in-depth research into complex regulatory networks and functional mechanisms.
MtoZ Biolabs provides, but is not limited to, the following protein post-translational modification analysis services:
Analysis Workflow
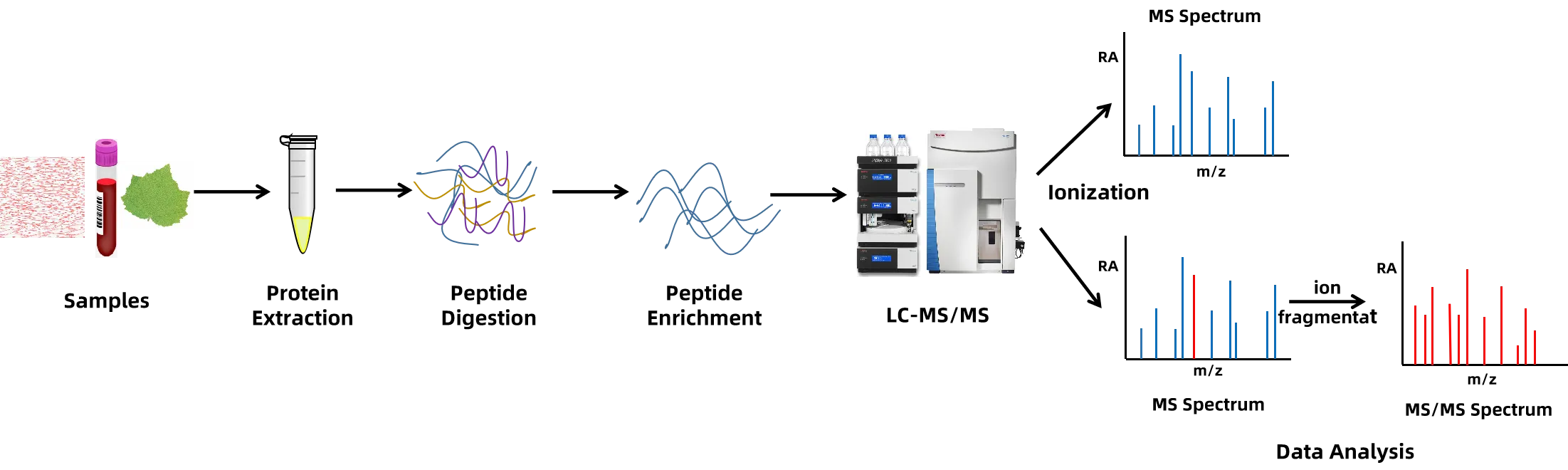
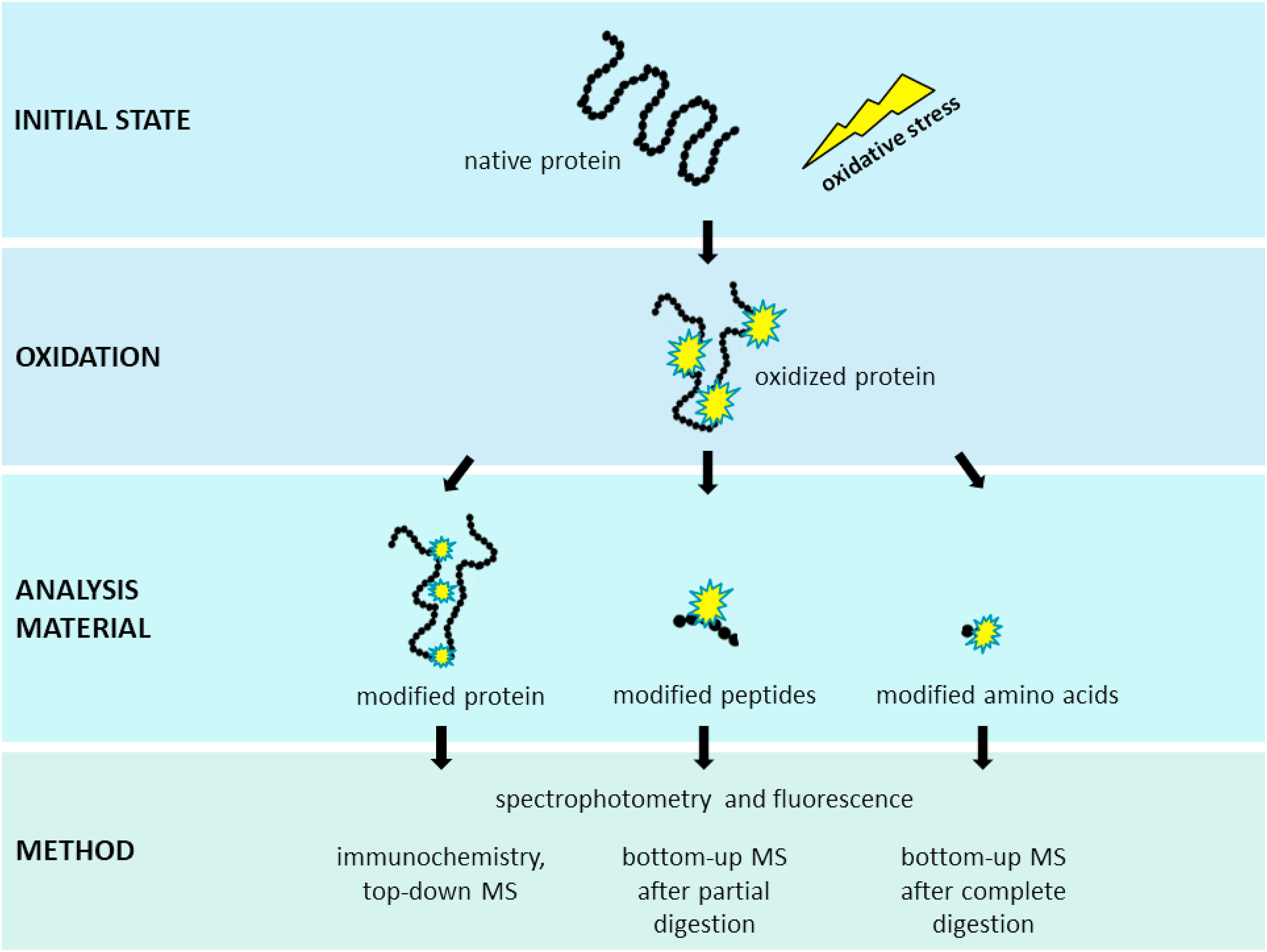
Figure 2. The Workflow of Protein Post-Translational Modifications Profiling.
Sample Submission Suggestions
1. Sample Type and Quantity
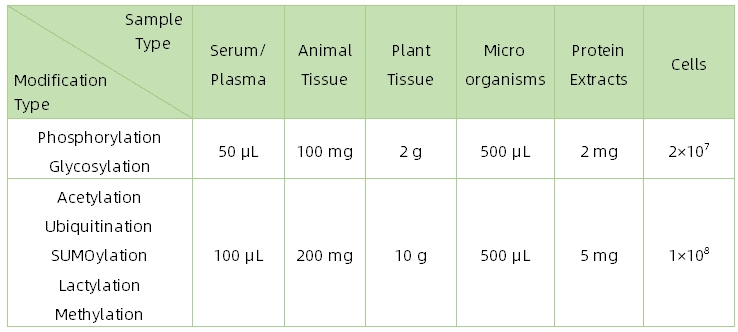
2. Sample Storage
Samples should be stored under low-temperature conditions (such as -80°C freezing) to avoid repeated freeze-thaw cycles, preventing protein degradation and loss of protein modifications.
3. Sample Transportation
During transportation, dry ice or cold-chain conditions should be used to ensure that the samples maintain stability and integrity before reaching the analytical platform.
Service Advantages
1. High-Resolution Detection
Based on an advanced LC-MS/MS platform, it enables highly sensitive identification of multiple PTM types.
2. Comprehensive Modification Coverage
Capable of detecting various modification types, including phosphorylation, acetylation, methylation, and ubiquitination.
3. Standardized Workflow
Optimized sample preparation and data analysis ensure accuracy and reproducibility of results.
4. Customized Solutions
Flexible analytical workflows are designed according to research objectives to meet diverse requirements.
Applications
✅Cell Biology: Enables analysis of cell cycle regulation, signal transduction, and stress response.
✅Molecular Genetics: Applicable to studies of gene expression regulation and epigenetic modifications.
✅Developmental Biology: Allows investigation of PTM dynamics across developmental stages or tissues.
✅Protein Function Research: Supports analysis of protein activity regulation, complex assembly, and interaction mechanisms.
✅Systems Biology: Facilitates integration of multi-omics data to construct protein modification regulatory network models.
FAQ
Q1: How Is the Accuracy of Site Identification Ensured?
A1: By utilizing a high-resolution LC-MS/MS platform combined with multi-stage fragmentation (MSn) and rigorous database matching algorithms, high-confidence localization and confirmation of modification types are achieved.
Q2: How Is the Detection of Low-Abundance Modifications Ensured?
A2: Optimized sample preparation and specific enrichment strategies (such as IMAC, TiO₂, and antibody-based enrichment) are employed to significantly enhance the detection rate of low-abundance modified peptides.
Q3: Does the Service Support Quantitative Analysis?
A3: Yes. Quantitative comparisons across multiple sample conditions can be performed using TMT/iTRAQ, Label-Free, or DIA strategies to obtain modification abundance variation trends.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Bioinformatics Analysis
5. Raw Data Files
Relying on advanced mass spectrometry platforms and a team of experienced specialists, MtoZ Biolabs provides comprehensive protein post-translational modifications profiling service for both scientific and industrial clients. From protein separation and enrichment to modification identification and data interpretation, we are committed to delivering reliable analytical support for your research. Contact us today to advance your exploration in proteomics and life sciences.