Histone Asymmetric Dimethylation Analysis Service
Based on the high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS) platform, the histone asymmetric dimethylation analysis service launched by MtoZ Biolabs enables systematic detection and quantitative analysis of asymmetric dimethylation (aDMA) modifications on arginine residues of histones. This service combines optimized sample preparation workflows with specific enrichment strategies to accurately identify aDMA modifications at different sites and evaluate their abundance changes, providing data on modification distribution, quantitative differences, and functional annotations to support studies on the biological functions of aDMA.
Overview
Histone asymmetric dimethylation (aDMA) is a post-translational modification catalyzed by arginine methyltransferases such as PRMT1 and PRMT3, in which two methyl groups are added to the same guanidino nitrogen of an arginine residue. This modification regulates interactions between histones and DNA or protein complexes, thereby influencing chromatin conformation and gene expression states. aDMA plays important roles in epigenetic regulation, signal transduction, and cellular differentiation, and it is widely applied in epigenetic research and molecular mechanism exploration.
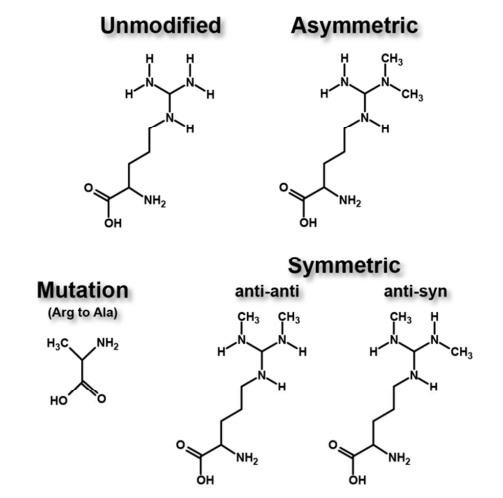
Li, Z H. et al. The Journal of Physical Chemistry B, 2018.
Figure 1. Modification Implemented on R42 in Histone H3.
Analysis Workflow
1. Sample Preparation
Histones are extracted from cells or tissues under mild conditions to preserve the integrity and stability of modifications.
2. Enzymatic Digestion
Proteins are digested under optimized conditions to generate peptides suitable for mass spectrometry detection.
3. Modified Peptide Enrichment
Specific enrichment strategies are employed to selectively capture asymmetrically dimethylated peptides.
4. Mass Spectrometry Detection
High-resolution LC-MS/MS is used to accurately identify and quantify aDMA modification sites.
5. Data Analysis
Bioinformatics tools are applied to analyze modification distribution and abundance differences, followed by report generation.
Sample Submission Suggestions
1. Sample Type and Quantity
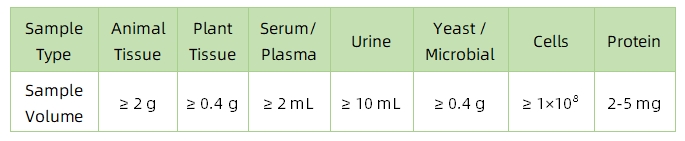
Note: Plasma should be collected using EDTA as an anticoagulant. Standard tissue or cell lysis buffers can be used during protein extraction.
2. Sample Transportation
Avoid repeated freeze-thaw cycles. Samples are recommended to be stored at -80°C and transported on dry ice to ensure low-temperature conditions throughout the process and prevent modification loss.
Note: For special samples or if a detailed submission plan is required, please contact MtoZ Biolabs technical staff in advance.
Service Advantages
1. High-Resolution Detection
Leveraging an advanced LC-MS/MS platform, this service enables precise identification and highly sensitive quantification of low-abundance aDMA modifications.
2. Specific Enrichment Strategy
An optimized peptide enrichment method is applied to significantly improve detection specificity and coverage.
3. Customized Experimental Design
Experimental workflows are flexibly tailored to different sample types and research objectives to meet diverse scientific needs.
4. Comprehensive One-Stop Service
End-to-end support is provided from sample preparation to final report delivery, streamlining the entire research process.
Applications
1. Epigenetic Regulation Research
The histone asymmetric dimethylation analysis service can be applied to investigate the role of aDMA in modulating chromatin structure and regulating gene expression.
2. Transcriptional Regulation Mechanism Exploration
By detecting aDMA modifications on key histone sites, this service helps elucidate their regulatory functions in transcriptional activation or repression.
3. Signal Pathway Studies
The histone asymmetric dimethylation analysis service enables the examination of dynamic changes in aDMA during cellular signaling and stress responses.
4. Potential Biomarker Discovery
By comparing aDMA modification patterns under different conditions, researchers can identify novel functional molecular biomarkers.
FAQ
Q1: Which Histone Sites Can Be Detected in Asymmetric Dimethylation Analysis?
A1: The service covers asymmetric dimethylation (aDMA) sites across multiple histone subtypes such as H3 and H4, including commonly studied sites like H3R2me2a and H4R3me2a. The detection range can also be extended based on specific research requirements.
Q2: What Is the Difference between aDMA and sDMA?
A2: aDMA refers to asymmetric dimethylation, where two methyl groups are attached to the same side nitrogen atom of an arginine residue, while sDMA (symmetric dimethylation) involves methylation on both nitrogen atoms. This structural difference leads to distinct roles in chromatin regulation and protein-protein interactions.







