Histone Lysine Methacrylation Analysis Service
Based on the high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) platform, the histone lysine methacrylation analysis service launched by MtoZ Biolabs enables precise detection and quantitative analysis of lysine methacrylation (Kmea) modifications on histones. By integrating efficient peptide enrichment and optimized sample preparation workflows, this service can identify Kmea modifications at different sites and analyze their abundance variations. Through the incorporation of bioinformatics tools, it provides data on modification distribution, quantitative differences, and functional annotation, supporting in-depth research on the biological functions of Kmea.
Overview
Histone lysine methacrylation (Kmea) is a novel lysine post-translational modification in which a methacryloyl group is covalently attached to the ε-amino group of lysine residues. This modification alters the charge and conformation of histones, thereby affecting chromatin structure and gene transcriptional regulation. Kmea is closely associated with cellular metabolism and energy homeostasis, playing important roles in chromatin remodeling, transcriptional regulation, and metabolic adaptation. Histone lysine methacrylation analysis is widely applied in epigenetic research, metabolic reprogramming mechanism studies, and potential biomarker discovery.
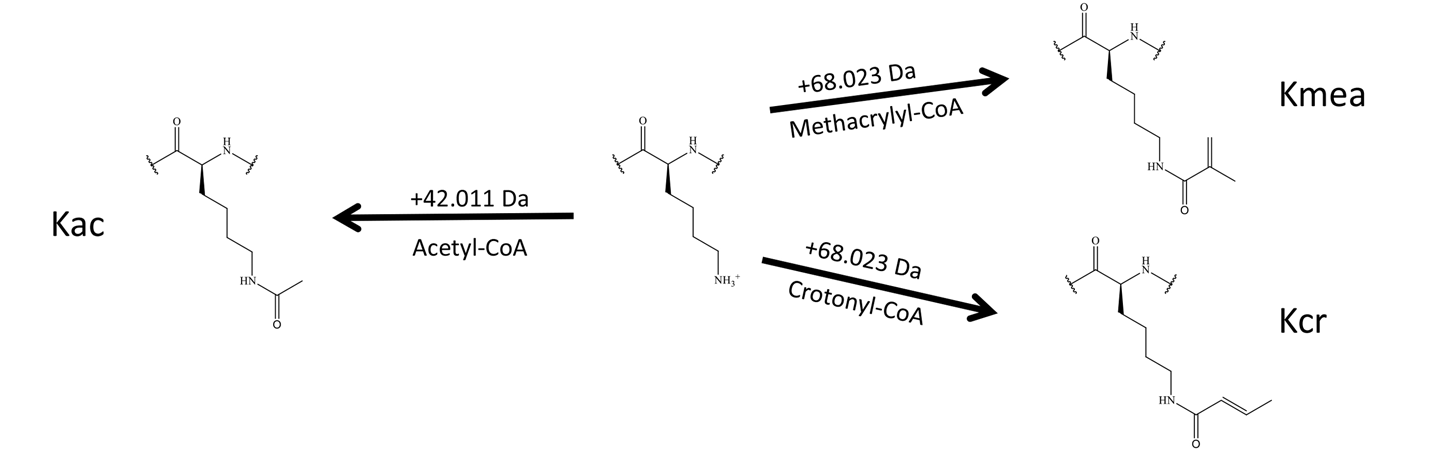
Delaney, K. et al. Cell Discovery, 2021.
Figure 1. Lysine (Center) Is Capable of Being Modified with Multiple Different PTMs.
Analysis Workflow
1. Histone Extraction and Digestion
Histones are isolated from the samples and enzymatically digested under optimized conditions to generate detectable peptides.
2. Modified Peptide Enrichment
A specific enrichment strategy is employed to selectively capture methacrylated peptides, improving detection sensitivity.
3. Mass Spectrometry Analysis
High-resolution LC-MS/MS is used to accurately identify and quantify Kmea modification sites.
4. Data Interpretation
Bioinformatics tools are integrated to provide comprehensive analysis of modification distribution and functional annotation results.
Sample Submission Suggestions
1. Sample Type and Quantity
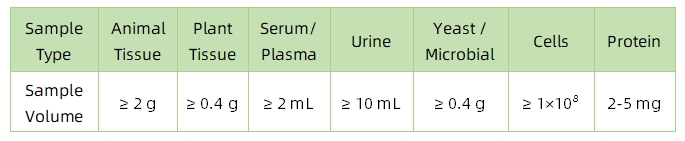
Note: Plasma should be collected using EDTA as an anticoagulant. Standard tissue or cell lysis buffers can be used during protein extraction.
2. Sample Transportation
Avoid repeated freeze-thaw cycles. Samples are recommended to be stored at -80°C and transported on dry ice to ensure low-temperature conditions throughout the process and prevent modification loss.
Note: For special samples or if a detailed submission plan is required, please contact MtoZ Biolabs technical staff in advance.
Service Advantages
1. High-Resolution Detection
Leveraging an advanced LC-MS/MS platform, this service enables precise identification of low-abundance lysine methacrylation modifications.
2. Optimized Enrichment Strategy
Employing efficient modified peptide enrichment technology significantly enhances detection sensitivity and site coverage.
3. Comprehensive Quality Control
Strict quality control is implemented throughout the entire workflow from sample preparation to data output to ensure reliability and reproducibility.
4. Flexible Experimental Design
Analysis workflows can be customized according to sample type and research objectives to meet diverse scientific needs.
Applications
1. Epigenetic Regulation Research
The histone lysine methacrylation analysis service can be applied to investigate the functional roles of Kmea modifications in chromatin conformation regulation and gene transcription.
2. Potential Biomarker Discovery
By comparing Kmea modification patterns under different experimental conditions, this service supports the identification of functional molecular biomarkers.
3. Cell Differentiation Research
The histone lysine methacrylation analysis service can be utilized to reveal the regulatory functions of Kmea during stem cell differentiation and developmental processes.
4. Integrative Multi-Omics Analysis
Combining transcriptomics, metabolomics, and other omics datasets enables a systematic exploration of Kmea-associated regulatory networks.
FAQ
Q1: What Distinguishes Methacrylation from Other Acylation Modifications?
A1: Lysine methacrylation (Kmea) contains a double-bond structure, which confers greater hydrophobicity and steric hindrance compared with other acyl modifications. This structural feature may exert a stronger impact on protein conformation and chromatin stability.
Q2: How Is the Specificity of Methacrylation Identification Ensured?
A2: High-resolution LC-MS/MS combined with precise mass filtering and characteristic fragment ion matching allows clear distinction between Kmea and structurally similar modifications (such as acetylation or propionylation). Specificity is further validated through database searching and manual verification.







