Protein Sulfation Analysis Service
Based on the high-resolution Orbitrap MS/MS platform combined with the nanoLC system, MtoZ Biolabs has launched the protein sulfation analysis service which enables systematic detection and quantification of sulfation modifications in protein samples. This service integrated with specific enrichment strategies and optimized experimental procedures, can comprehensively analyze the distribution, modification levels, and dynamic changes of sulfation sites, as well as provide functional annotation of modification features. The final output data not only include accurate identification results and quantitative variation information of sulfation modifications but also offer reliable support for researchers in signal pathway studies, cell communication, and structural-functional analysis.
Overview
Protein sulfation is defined as the process in which sulfate groups are covalently attached to tyrosine or other amino acid residues of proteins through enzymatic reactions catalyzed by sulfotransferases. This modification can significantly alter the charge state and spatial conformation of proteins, thereby affecting their interactions with receptors, ligands, and other molecules. Protein sulfation analysis has been widely applied in signal transduction studies, cell communication mechanism analysis, immunology research, and structural biology, providing important data support for elucidating protein function regulation and complex biological networks.
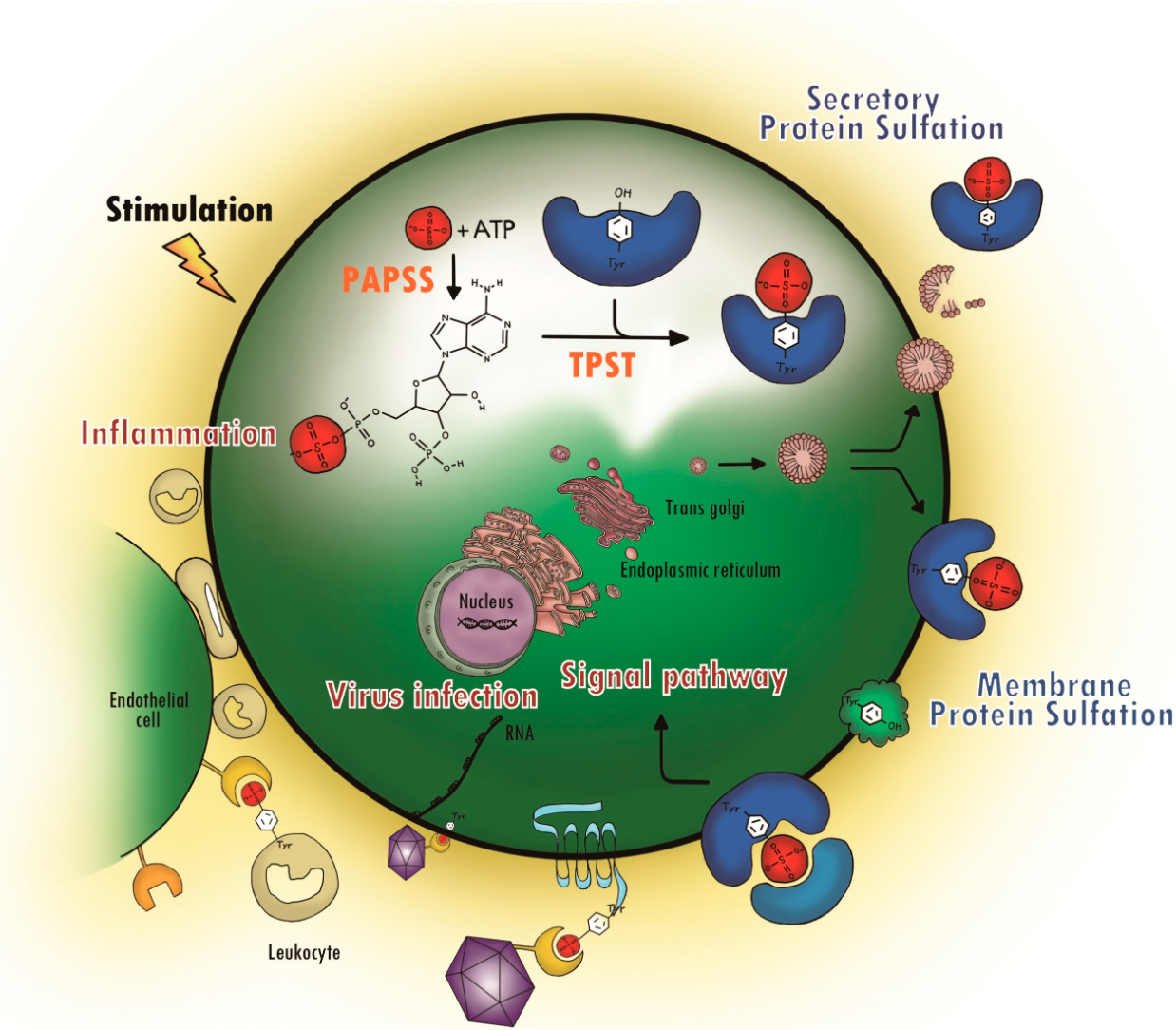
Yang, Y S. et al. Molecules, 2015.
Figure 1. Protein Tyrosine Sulfation (PTS) and Its Biological Path.
Services at MtoZ Biolabs
1. Target Protein Sulfation Analysis
MtoZ Biolabs can perform site-specific sulfation detection for designated target proteins, identifying tyrosine or other residues that may undergo sulfation. Relying on a high-resolution LC-MS/MS platform, we are able to compare sulfation changes under different treatment conditions, providing accurate evidence for studying protein functional regulation and signaling pathways.
2. Sulfated Proteomics Analysis
By combining enrichment strategies with high-throughput mass spectrometry, MtoZ Biolabs can systematically identify and quantify sulfated peptides in complex systems, constructing a global sulfation profile. This service can be used to elucidate the distribution characteristics and functional patterns of sulfation in cell communication, receptor recognition, and disease-related regulation.
Analysis Workflow
1. Sample Preparation
Extract target proteins from cells, tissues, body fluids, or purified protein samples, and perform quantitative detection to ensure uniform quality.
2. Protein Digestion
Digest proteins into peptides using trypsin or other enzymes to facilitate the detection and localization of sulfation modifications.
3. Sulfated Peptide Enrichment
According to the complexity of the sample and research objectives, apply specific antibodies or chemical capture methods to selectively enrich sulfated peptides.
4. Mass Spectrometry Detection
Analyze enriched peptides using the nanoLC system coupled with the Orbitrap MS/MS platform. ETD or EThcD fragmentation techniques can be employed to preserve unstable sulfate groups.
5. Data Analysis
Combine database searches and bioinformatics tools to annotate sulfation modification sites, providing distribution characteristics, modification levels, and functional interpretation.
Service Advantages
1. High-Sensitivity Detection
Relying on a high-resolution LC-MS/MS platform, it can capture low-abundance sulfation modifications in complex sample backgrounds, ensuring the accuracy and reliability of detection results.
2. Specific Enrichment
By combining antibodies or chemical capture methods, sulfated peptides can be effectively separated, improving the signal-to-noise ratio and the detection rate of modification sites.
3. One-Stop Service
From sample preparation and modification detection to data analysis, a complete workflow and technical support are provided to help researchers efficiently obtain systematic data.
4. Customized Solutions
MtoZ Biolabs can flexibly customize experimental plans according to your research objectives and samples to meet individualized experimental needs.
Applications
1. Signal Transduction Research
By detecting sulfation modification sites, their regulatory roles in key signaling pathways can be revealed, helping to analyze intracellular and extracellular signal transmission mechanisms.
2. Cell Communication Analysis
Protein sulfation analysis service can be used to study the impact of sulfation modifications on ligand–receptor binding and molecular recognition in cell-to-cell interactions.
3. Immunology Research
By detecting sulfation modifications of immune-related proteins, their roles in immune recognition and immune regulation can be explored.
4. Environmental Stress Response
Protein sulfation analysis service can be used to monitor changes in sulfation modifications under external environmental or stress conditions, analyzing mechanisms of cellular adaptation and homeostasis regulation.
FAQ
Q1: Is Sulfation Modification Stable in Mass Spectrometry Detection?
A1: Some sulfation modifications are prone to signal loss during ionization and fragmentation. Therefore, optimized fragmentation modes (such as ETD or EThcD) are usually applied to improve the retention of modification information and detection reliability.
Q2: Can Sulfation Be Quantitatively Analyzed?
A2: Yes. Common strategies include label-free quantification, isotopic labeling (TMT, iTRAQ), and DIA analysis, which can be selected according to the experimental objectives.
Q3: What Precautions Should Be Taken for Sample Submission?
A3: It is recommended to provide sufficient amounts of cells, tissues, or purified protein samples, and to store them at -80°C to avoid repeated freeze-thaw cycles. During transportation, dry ice or cold chain conditions should be used to ensure that the modification state is not compromised.







