Quantitative Histone PTMs Analysis Service
Based on the high-resolution LC-MS/MS platform and stable quantitative systems, the quantitative histone PTMs analysis service launched by MtoZ Biolabs enables simultaneous detection and quantitative evaluation of multiple histone post-translational modifications (PTMs). This service supports quantitative strategies such as label-free, TMT/iTRAQ, and DIA/SWATH, allowing precise measurement of relative abundances and dynamic changes of different modification types. It provides site-specific abundance, differential statistics, and pathway correlation results, offering reliable quantitative evidence for epigenetic regulation research.
Overview
Histone post-translational modifications (PTMs) refer to chemical modifications that occur on histones after translation, such as methylation, acetylation, and phosphorylation. These modifications finely regulate epigenetic processes by influencing chromatin structure and gene transcriptional activity. Since the dynamic variation in types and abundances of modifications directly determines gene expression states, quantitative analysis is crucial for revealing the strength and biological function of regulatory mechanisms. Quantitative histone PTMs analysis helps researchers accurately assess modification level changes, elucidate chromatin regulation mechanisms, and provide key data support for studies in epigenetics, cellular signaling, and developmental biology.
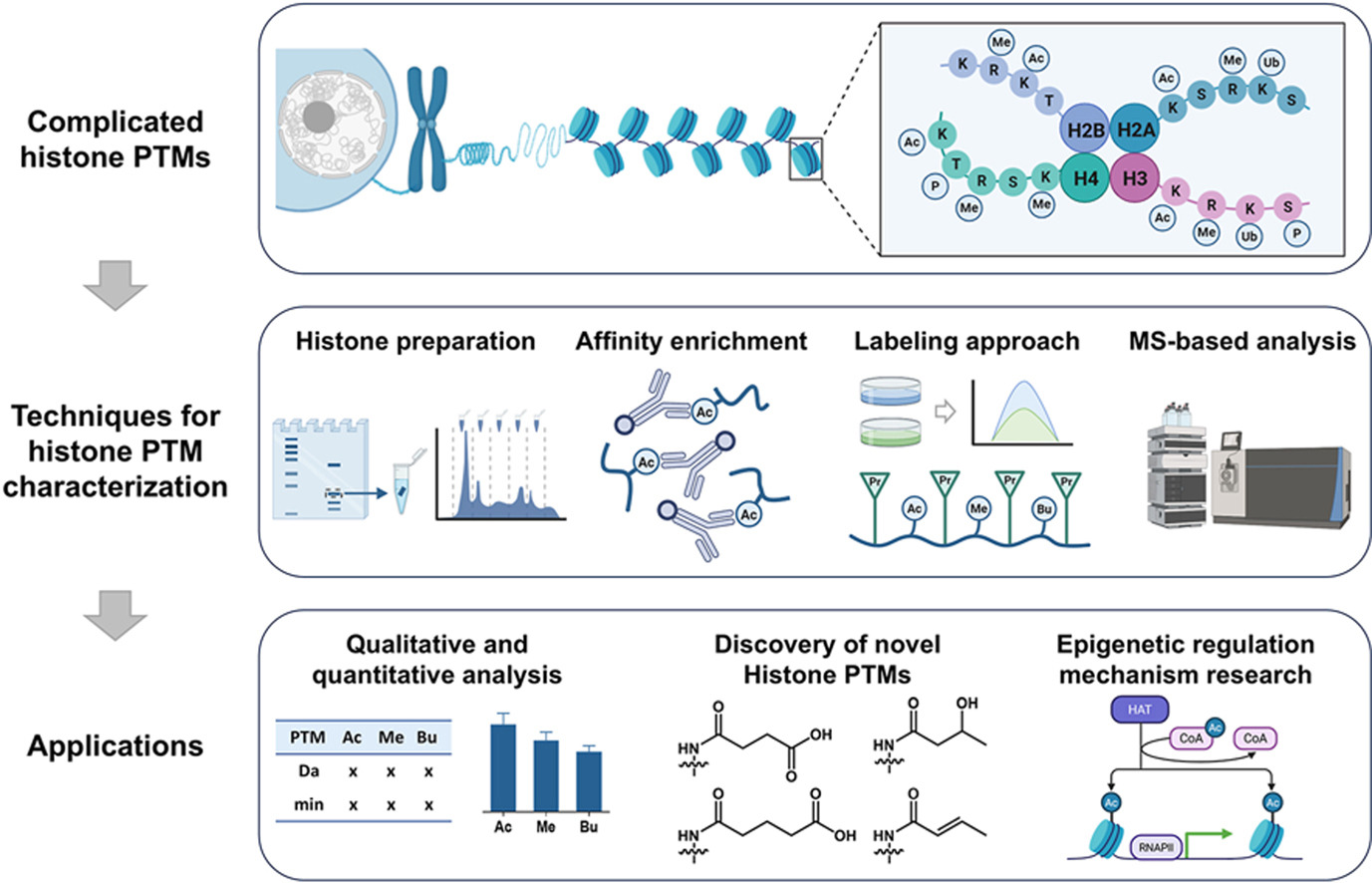
Zhao, W S. et al. Current Opinion in Chemical Biology, 2025.
Figure 1. MS-based Characterization of Histone Post-Translational Modification.
MS-based Methods for Histone PTMs Quantification
MtoZ Biolabs has developed different methods for quantification of histone PTM profiles, including but not limited to the following:
1. SILAC Quantification
By incorporating stable isotope-labeled amino acids into cell culture, this approach enables precise quantification of histone post-translational modifications under different conditions, making it suitable for dynamic studies at the cellular level.
2. Label-Free Quantification
Without the need for labeling, histone modifications are quantified by comparing peptide signal intensities or peak areas. This method is simple to operate and well-suited for large-scale sample analysis.
3. TMT/iTRAQ Quantification
Using isobaric tags for chemical labeling of multiple sample groups, this strategy achieves multiplexed detection. Quantification is based on reporter ion signals, offering high sensitivity and reproducibility in histone modification analysis.
4. DIA/SWATH Quantification
Employing data-independent acquisition (DIA) or SWATH-MS, this unbiased approach combines spectral libraries to achieve high coverage and full-spectrum quantification of histone modifications, ideal for complex samples and multi-PTM integrative studies.
Analysis Workflow
1. Sample Preparation
Histones are extracted from samples, followed by enzymatic digestion and enrichment of modified peptides.
2. LC-MS/MS Detection
High-resolution mass spectrometry is used to achieve precise identification and quantitative analysis of histone modifications.
3. Data Processing and Quantitative Analysis
Spectral identification, modification site localization, and abundance calculation are performed using specialized software to generate quantitative results.
4. Bioinformatics Interpretation
Functional and pathway enrichment analyses are conducted to reveal the regulatory roles of histone modifications.
Sample Submission Suggestions
1. Sample Type and Quantity
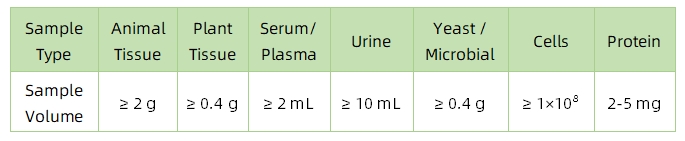
Note: Plasma should be collected using EDTA as an anticoagulant. Standard tissue or cell lysis buffers can be used during protein extraction.
2. Sample Transportation
Avoid repeated freeze-thaw cycles. Samples are recommended to be stored at -80°C and transported on dry ice to ensure low-temperature conditions throughout the process and prevent modification loss.
Note: For special samples or if a detailed submission plan is required, please contact MtoZ Biolabs technical staff in advance.
Service Advantages
1. High-Resolution Quantitative Capability
Leveraging advanced LC-MS/MS platforms, this service enables high-sensitivity, wide dynamic range quantification of diverse histone modifications.
2. Multi-Strategy Quantitative System
Supports multiple quantitative methods, including SILAC, TMT/iTRAQ, Label-Free, and DIA approaches, to accommodate different experimental requirements.
3. Efficient PTM Enrichment Technology
Employs optimized modified peptide enrichment protocols to significantly enhance the detection rate of low-abundance PTMs.
4. Comprehensive One-Stop Service
Provides a complete workflow covering sample preparation, mass spectrometry detection, and data reporting.
Applications
1. Epigenetic Regulation Research
Quantitative histone PTMs analysis service can be applied to evaluate dynamic changes of histone modifications involved in chromatin structure and transcriptional regulation.
2. Cell Differentiation and Development Studies
By comparing modification abundance across developmental stages, this service helps reveal the regulatory roles of histone PTMs in cell fate determination.
3. Metabolic Regulation Research
Quantitative histone PTMs analysis service enables the exploration of how cellular metabolic states influence histone modification patterns.
4. Functional Biomarker Discovery
Quantitative analysis of PTM differences facilitates the identification of potential key regulatory sites or functional biomarkers.







