Immunoaffinity Precipitation-Based Modified Peptide Enrichment Service
Post-translational modifications (PTMs) are key molecular events that regulate protein activity, localization, and interactions. Detecting these modifications accurately requires sensitive and specific enrichment techniques capable of isolating low-abundance modified peptides from complex proteomes. Immunoaffinity precipitation-based enrichment has emerged as one of the most precise and targeted strategies for studying PTMs, offering unparalleled specificity through antibody–antigen interactions.
MtoZ Biolabs provides a professional Immunoaffinity Precipitation-Based Modified Peptide Enrichment Service to support researchers in capturing and quantifying specific PTMs with high sensitivity and reliability. Our service integrates high-quality, modification-specific antibodies with optimized immunoprecipitation workflows and advanced LC-MS/MS detection. By combining antibody-based selectivity with the resolution of modern mass spectrometry, we deliver robust datasets that enable comprehensive characterization of PTM landscapes. We also offer multiplexed IP-MS, co-immunoprecipitation (Co-IP), and hybrid enrichment approaches that can be tailored for phosphorylation, ubiquitination, acetylation, methylation, SUMOylation, and other PTM analyses.
Technical Principles
Immunoaffinity precipitation is an antibody-driven approach that selectively isolates proteins or peptides carrying specific modifications. The technique relies on high-affinity binding between antibodies and epitopes containing distinct PTM signatures, such as phosphoserine, acetyl-lysine, or ubiquitin-conjugated residues. By conjugating antibodies to solid-phase supports (e.g., agarose or magnetic beads), modified targets can be efficiently captured, washed, and eluted under mild conditions to preserve modification integrity.
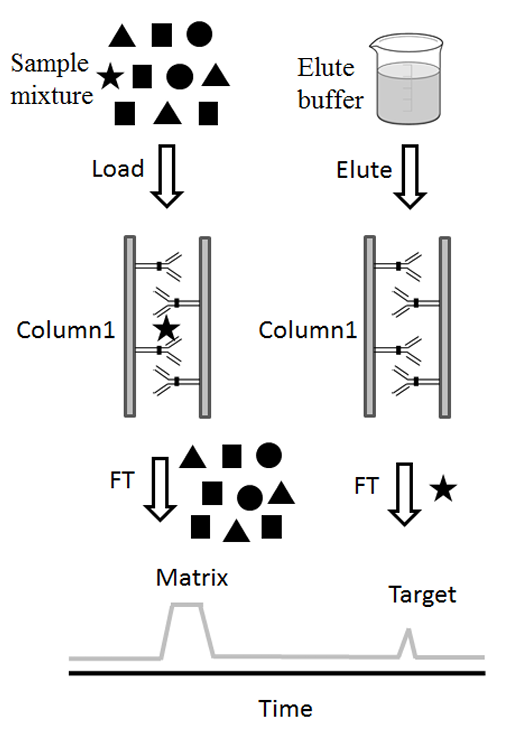
Wu, C. et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2016.
Figure 1. The Main Workflows of Immunoenrichment.
(The Star Represent the Target Molecules and Other Symbols are Sample Matrix.)
Analysis Workflow
1. Sample Preparation
Proteins are extracted from cells, tissues, or biofluids, quantified, and digested into peptides while ensuring preservation of PTMs.
2. Antibody Coupling and Immunoprecipitation
Modification-specific antibodies are immobilized on magnetic or agarose beads, followed by incubation with the sample to capture modified peptides or proteins.
3. Washing and Elution
Non-specifically bound peptides are removed, and the bound modified peptides are gently eluted under optimized conditions to maintain modification stability.
4. Mass Spectrometry Analysis
The enriched peptides are analyzed using high-resolution LC-MS/MS, enabling site-specific identification, quantification, and structural confirmation of PTMs.
5. Data Analysis and Bioinformatics
Comprehensive bioinformatics pipelines are applied to annotate PTM sites, quantify modification changes, and map associated signaling pathways or protein interaction networks.
Sample Submission Suggestions
1. Sample Type: Cell lysates, tissue homogenates, serum/plasma, or purified proteins.
2. Amount Required: Minimum 500 µg total protein per sample.
3. Storage and Transport: Samples should be snap-frozen in liquid nitrogen and shipped on dry ice.
*Note: For complex or low-yield samples, contact MtoZ Biolabs for tailored pre-processing guidance.
Service Advantages
✅ High Specificity and Sensitivity
Modification-specific antibodies ensure selective capture of target peptides, minimizing background noise.
✅ Comprehensive Analytical Coverage
Compatible with phosphorylation, acetylation, ubiquitination, methylation, and other PTMs for both protein- and peptide-level analysis.
✅ Customizable Experimental Design
Tailored immunoprecipitation strategies for targeted or global studies based on project goals and sample availability.
✅ Expert Technical Support
Our experienced proteomics team provides data interpretation, troubleshooting, and optimization guidance throughout the project.
Applications
1. Signal Transduction Studies
Investigate phosphorylation, ubiquitination, and SUMOylation pathways to understand dynamic signaling regulation.
2. Epigenetic Research
Profile histone acetylation and methylation patterns to uncover epigenetic mechanisms controlling gene expression.
3. Disease Mechanism Exploration
Identify aberrant PTM signatures associated with cancer, neurodegeneration, inflammation, and autoimmune disorders.
4. Drug Target Validation
Evaluate how pharmacological interventions alter PTM levels or signaling networks in preclinical models.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
By combining high-quality antibody-based enrichment with advanced mass spectrometry and bioinformatics, MtoZ Biolabs enables researchers to detect, quantify, and interpret modification events with confidence. Free project evaluation, welcome to learn more details!







