mRNA Modification LC-MS Analysis Service
Based on a high-resolution LC-MS/MS platform, the mRNA modification LC-MS analysis service launched by MtoZ Biolabs enables precise detection and quantitative analysis of chemical modifications within mRNA molecules. Through nucleotide extraction, enzymatic digestion, and high-resolution mass spectrometric measurement, this service systematically identifies the distribution characteristics and variation trends of modifications. Combined with standard calibration and database matching, it provides high-confidence modification profiling data to support studies on RNA modification mechanisms, gene regulation analysis, and post-transcriptional functional characterization.
Overview
mRNA modification refers to various chemical modifications occurring on messenger RNA molecules, such as methylation (m6A, m1A), pseudouridylation (Ψ), and 2′-O-methylation. These modifications play crucial roles in regulating mRNA stability, splicing, translation efficiency, and degradation, serving as key components of epitranscriptomic research. The dynamic changes of mRNA modifications are closely associated with gene expression regulation, cell differentiation, stress response, and disease progression. Research in this field has been widely applied to biology, medicine, drug development, and biomarker discovery, providing essential theoretical and technical support for understanding RNA functional regulation.
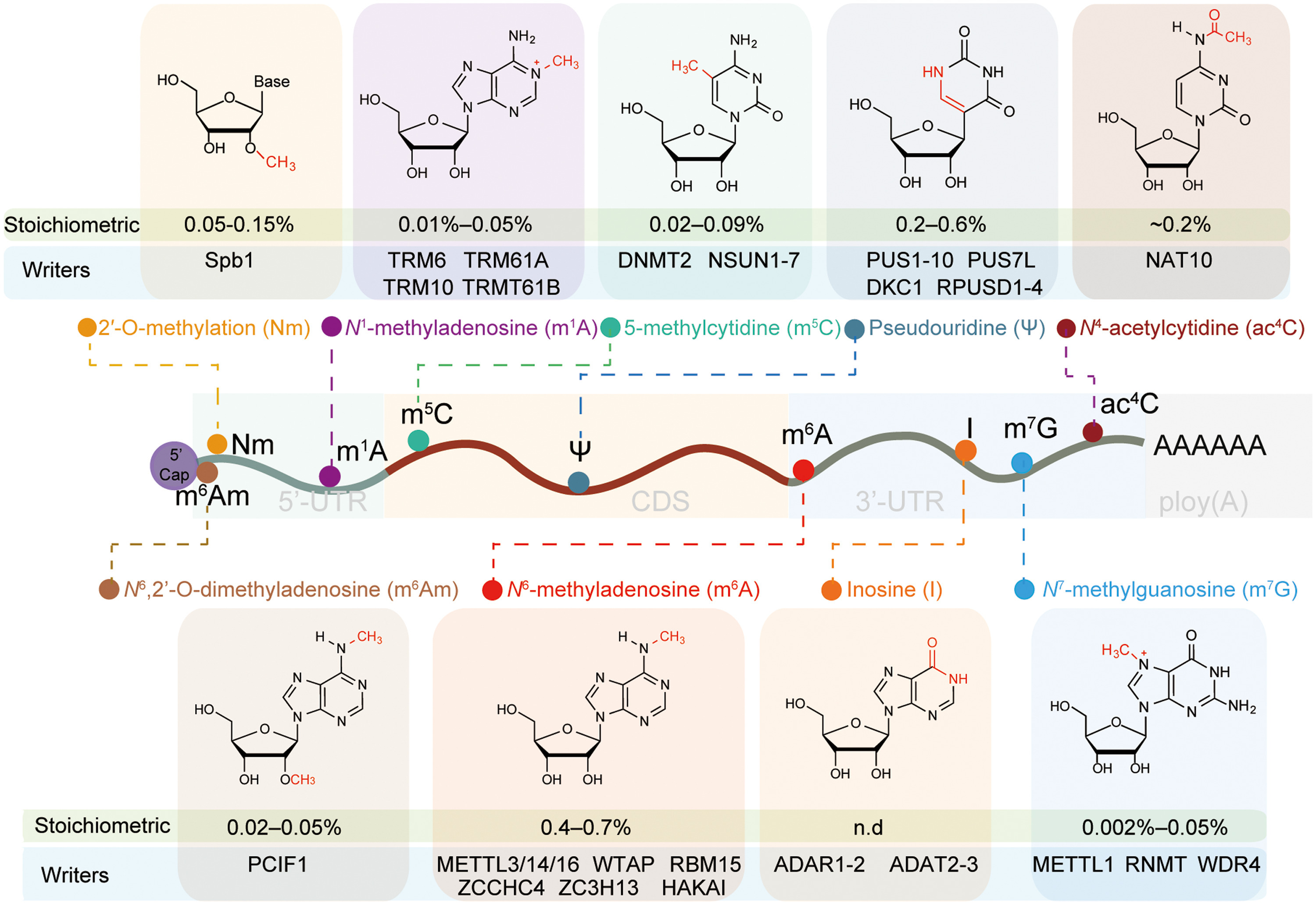
Li, K. et al. Fundamental Research, 2023.
Figure 1. Distribution, Chemical Structures, and Regulatory Proteins of mRNA Modifications.
Analysis Workflow
1. Sample Preparation and RNA Extraction
Total RNA is extracted from the sample, followed by mRNA isolation and purification.
2. Enzymatic Digestion and Nucleoside Generation
The purified mRNA is enzymatically digested into individual nucleosides.
3. LC-MS/MS Detection
High-resolution liquid chromatography–mass spectrometry (LC-MS/MS) is used to analyze the nucleoside samples.
4. Data Interpretation and Quantitative Analysis
By combining standard calibration and database searching, modification types are identified, and relative abundances are calculated to generate modification profiles.
5. Result Integration and Report Delivery
Detection and quantification results are integrated to provide a standardized report including modification distribution, abundance analysis, and visual data presentation.
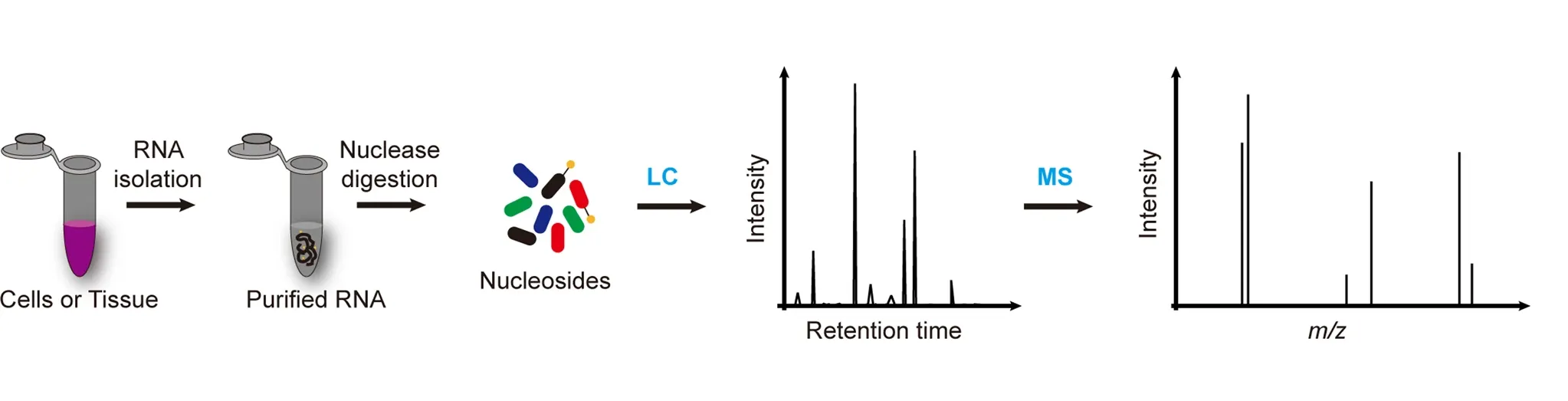
Zhang Y. et al. Exp Mol Med. 2022.
Figure 2. The Workflow of mRNA Modification LC-MS Analysis.
Sample Submission Suggestions
1. Sample Type
Cell, tissue, blood, and culture samples are accepted, as well as total RNA or purified mRNA forms.
Note: Samples should be free from degradation and contamination to ensure analytical accuracy.
2. Sample Storage
RNA samples should be stored in an RNase-free environment at -80°C for long-term preservation or at -20°C for short-term storage. Avoid repeated freeze–thaw cycles and exposure to high temperatures.
3. Sample Transportation
Liquid samples should be shipped on dry ice via a cold chain, while lyophilized samples may be sent at ambient temperature for short periods. All samples must be sealed and protected from light to prevent RNA degradation and loss of modifications.
Service Advantages
1. High-Resolution Detection Platform
Utilizing an advanced LC-MS/MS system, this service enables highly sensitive detection and precise quantification of mRNA modifications.
2. Comprehensive Modification Profiling
Capable of identifying multiple types of mRNA chemical modifications simultaneously to reveal their distribution characteristics and dynamic changes.
3. One-Stop Research Support
Provides end-to-end service covering RNA extraction, mass spectrometry detection, and data interpretation.
4. Customized Analytical Solutions
Experimental strategies can be flexibly designed according to specific research goals to meet diverse analytical needs.
Applications
1. Post-Transcriptional Regulation Studies
The mRNA modification LC-MS analysis service can be used to elucidate the roles of various modifications in mRNA stability, nuclear export, and translational regulation.
2. Development and Differentiation Research
By analyzing changes in mRNA modifications during developmental stages or cell differentiation, this service helps reveal their spatiotemporal characteristics in gene regulation.
3. RNA Metabolism and Modification Dynamics
The mRNA modification LC-MS analysis service enables monitoring of RNA modification formation, removal, and conversion processes to assess their dynamic balance and regulatory mechanisms.
4. Stress Response and Environmental Adaptation
By examining modification changes under heat, oxidative, nutrient, or pharmacological stress conditions, it supports exploration of cellular adaptive and regulatory pathways.
FAQ
Q1: Does the Purity of mRNA Samples Affect the Detection Results?
A1: Yes. High-purity mRNA samples significantly improve detection sensitivity and quantitative accuracy. It is recommended to use purified samples with rRNA and tRNA removed.
Q2: What Factors May Cause Analytical Errors?
A2: RNA degradation, residual salts, metal ions, or organic solvents can interfere with signal detection. Therefore, samples should be thoroughly purified and maintained at low ionic strength.
Q3: Can Modification Data Be Integrated with Transcriptomic or Proteomic Data?
A3: Yes. The results can be integrated with transcriptomic or proteomic datasets to reveal the influence of RNA modifications on transcriptional regulation and translation processes.







