N-Terminal PTM Analysis Service
Based on the high-resolution mass spectrometry (LC-MS/MS) platform and liquid separation system, the N-terminal PTM analysis service launched by MtoZ Biolabs can perform systematic detection and quantification of post-translational modifications at the N-terminus of proteins. This service, through specific modified peptide enrichment and optimized sample processing procedures, can accurately identify modification sites and analyze their distribution characteristics and modification abundance. The final output data include the identification results of modification sites, quantitative differences in modification levels, and related functional annotations, providing reliable support for researchers in protein function regulation, signal pathway analysis, and cell biology research.
Overview
N-terminal post-translational modifications (N-terminal PTMs) refer to chemical or enzymatic modifications occurring at the amino terminus of proteins, including acetylation, formylation, methylation, deformylation, and other forms. These modifications play important roles in protein stability, subcellular localization, degradation pathways, and intermolecular interactions. N-terminal post-translational modification analysis can help researchers explore protein degradation signals (N-end rule pathway), signal pathway regulation, and regulatory mechanisms in epigenetic processes, while also being widely applied in drug development, biomarker screening, and protein function research, providing reliable support for a deeper understanding of protein dynamic regulation.
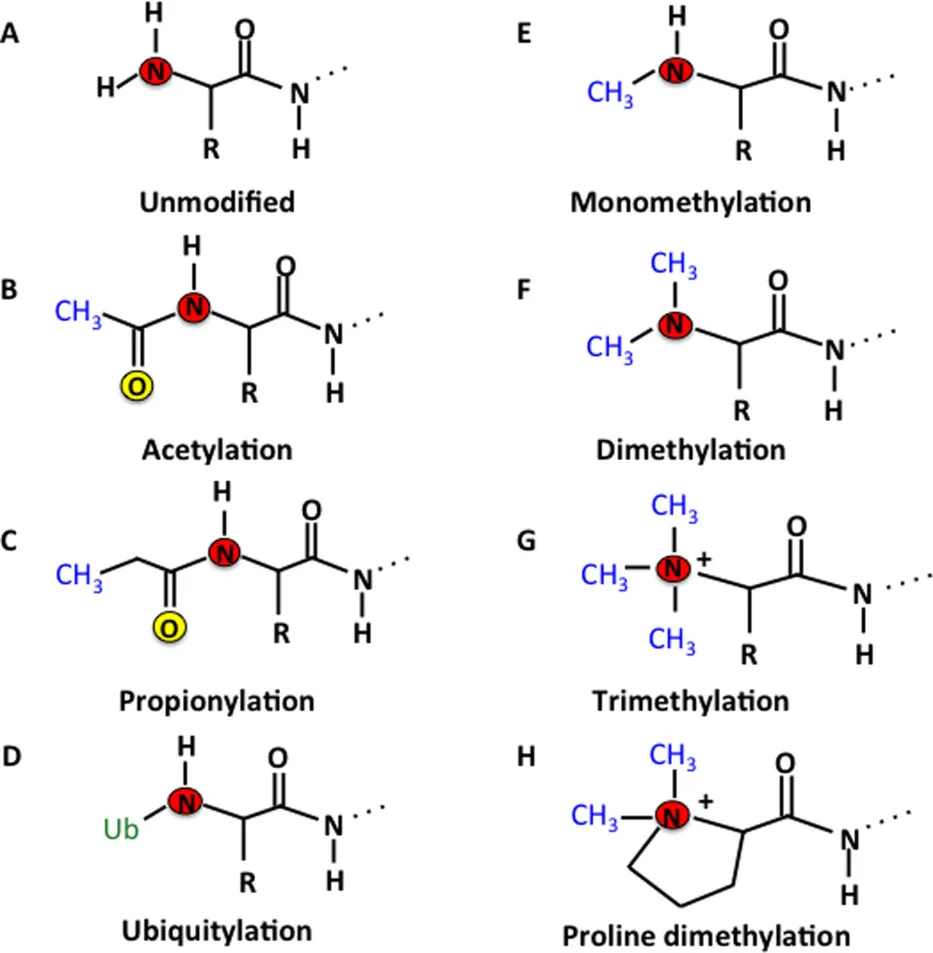
Tooley, J G. et al. Protein Science, 2014.
Figure 1. Various N-Terminal Post-Translation Modification Types.
Services at MtoZ Biolabs
1. Target Protein N-terminal PTM Analysis
MtoZ Biolabs can perform precise identification of N-terminal modification types and modification sites for specific target proteins, including acetylation, formylation and other specific modifications. By integrating high-resolution LC-MS/MS, we can analyze N-terminal structural states and compare modification differences under different conditions.
2. Proteomics N-terminal PTM Analysis
Relying on enrichment strategies and large-scale mass spectrometry platforms, MtoZ Biolabs can systematically obtain global N-terminal peptide information and identify N-terminal modification features across multiple proteins. This analysis enables rapid construction of N-terminal modification maps, helps reveal global regulatory patterns, and supports in-depth studies of protein processing and functional mechanisms.
Analysis Workflow
1. Sample Preparation
Extract proteins from the target samples and perform necessary preprocessing to ensure detection stability.
2. Protein Digestion
Perform enzymatic digestion under optimized conditions to generate peptides suitable for mass spectrometry analysis.
3. Modified Peptide Enrichment
Enrich N-terminal modified peptides through specific methods.
4. Mass Spectrometry Detection
Rely on high-resolution LC-MS/MS combined with liquid separation to perform accurate qualitative and quantitative analysis of modified peptides.
5. Data Analysis
Use databases and bioinformatics tools to output modification site distribution, modification levels, and functional annotations.
Sample Submission Suggestions
1. Sample Types
Various types of samples, such as cells, tissues, biofluids, and purified proteins, are acceptable. Samples must contain sufficient protein amounts to ensure reliable detection of modifications.
2. Sample Storage
It is recommended to store samples at -80°C for long-term preservation and to avoid repeated freeze-thaw cycles to minimize protein degradation and N-terminal modification loss.
3. Sample Transportation
Samples should be transported on dry ice or under cold-chain conditions, using sealed containers to ensure sample integrity and modification stability during transportation.
Service Advantages
1. High-Resolution Detection
Relying on the advanced LC-MS/MS platform, accurately capture low-abundance N-terminal modifications in complex samples to ensure reliable results.
2. Multiple Modification Coverage
Support systematic detection and quantification of various N-terminal PTMs, providing comprehensive and complete data output.
3. One-Stop Workflow
Cover the entire process from sample preparation, modification detection, to data analysis, providing researchers with complete solutions.
4. Customized Solutions
Adjust analytical strategies flexibly according to experimental objectives and sample characteristics to meet diverse research needs.
Applications
1. Signal Pathway Research
The N-terminal PTM analysis service can be used to reveal its regulatory role in key signaling pathways and support the analysis of cellular signaling networks.
2. Protein Degradation Mechanism Research
Exploring the role of N-terminal modifications in protein degradation pathways helps to analyze protein stability and metabolic regulation.
3. Cellular Function Analysis
By monitoring the dynamic changes of N-terminal modifications under different conditions, it clarifies their functions in cells.
4. Biomarker Discovery
The N-terminal PTM analysis service can be used to screen potential functional or state-related biomarkers, providing references for disease research and molecular diagnostics.
FAQ
Q1: Can Different Modifications Be Detected Simultaneously?
A1: Relying on the high-resolution mass spectrometry platform, parallel detection of multiple N-terminal modifications can be achieved, but complex samples may require stepwise enrichment.
Q2: Can the Detected Modifications Be Quantified?
A2: Yes. Common quantitative strategies include label-free quantification, isotope labeling (such as TMT, iTRAQ), and DIA. Appropriate methods can be selected flexibly according to experimental requirements.
Q3: What Are the Main Difficulties in Analysis?
A3: N-terminal modifications are often low in abundance and unstable, and some modifications lack complete database annotations, which may increase identification difficulty and require experimental validation.







