m6A Modification LC-MS Analysis Service
Based on a high-resolution LC-MS/MS platform, MtoZ Biolabs has launched the m6A modification LC-MS analysis service which enables precise detection and quantitative analysis of N6-methyladenosine (m6A) modifications in mRNA molecules. By integrating RNA extraction, enzymatic digestion, and high-resolution mass spectrometry detection, this service systematically characterizes the types, distribution, and abundance changes of m6A modifications. Through standard calibration and database comparison, high-confidence modification profiles can be obtained, providing researchers with reliable data support.
What is m6A modification?
m6A modification is one of the most abundant and extensively studied epitranscriptomic modifications found in messenger RNA (mRNA) and various noncoding RNAs. It regulates RNA stability, splicing, nuclear export, and translation efficiency by adding a methyl group at the N6 position of adenosine. Controlled by methyltransferase complexes ("writers"), demethylases ("erasers"), and binding proteins ("readers"), m6A plays a critical role in gene expression regulation networks. It is widely applied in studies of post-transcriptional regulation, developmental biology, RNA metabolism, cell differentiation, and epigenetics.
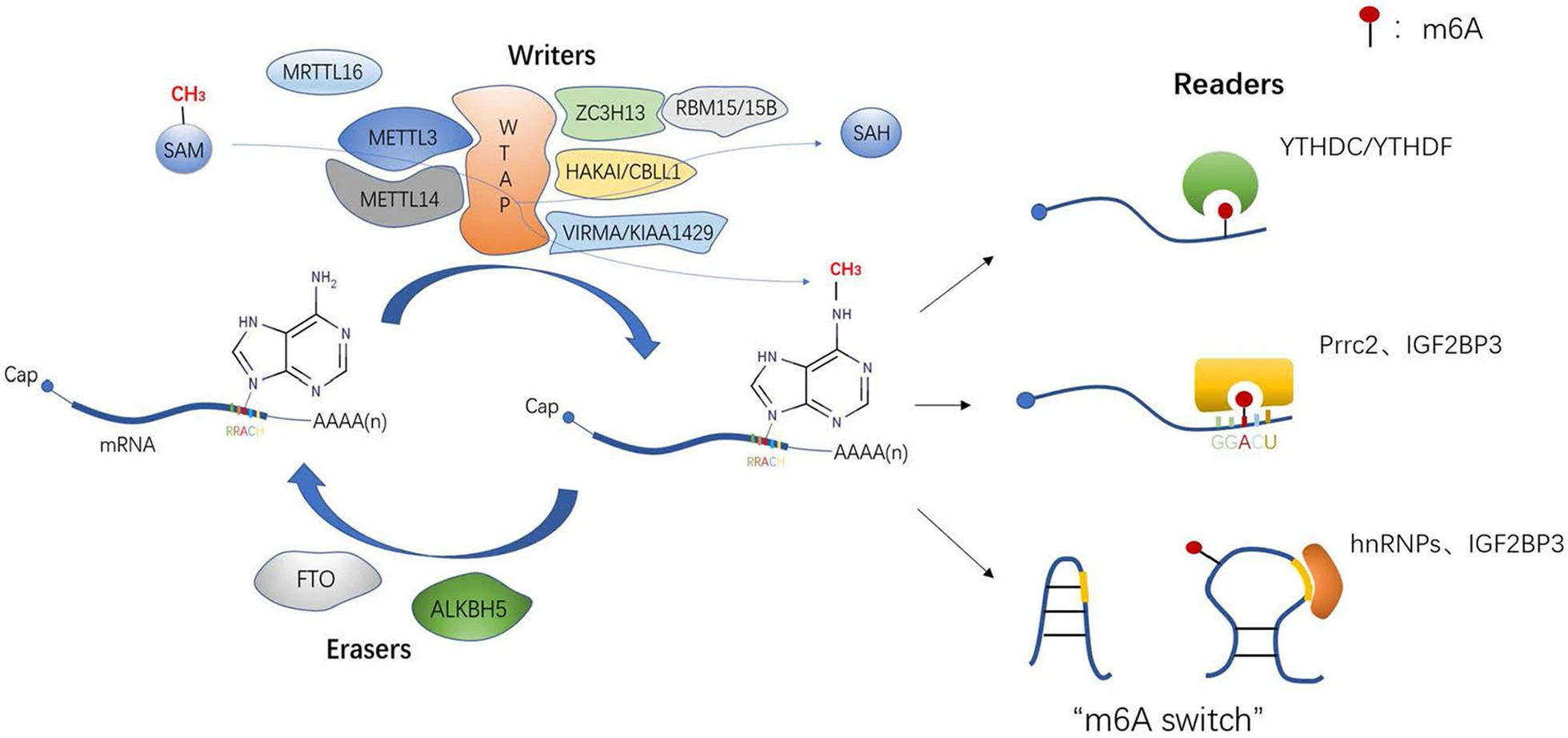
Wang, S S. et al. Cancer Cell International, 2022.
Figure 1. Dynamic and Reversible Process of m6A Modification and Three Recognition Methods of Readers.
Analysis Workflow
1. RNA Extraction and Purification
Total RNA is extracted from the samples, and high-purity mRNA is obtained after the removal of rRNA and tRNA.
2. Enzymatic Digestion and Nucleoside Preparation
The mRNA sample is enzymatically digested to generate individual nucleosides for downstream analysis.
3. Mass Spectrometry Detection and Signal Acquisition
A high-resolution LC-MS/MS system is used to separate and detect modified nucleoside signals, generating highly sensitive spectral data.
4. Data Processing and Modification Identification
By comparing with standard references and performing database analysis, the presence and abundance variation of m6A modifications are identified.
5. Result Integration and Report Generation
Modification data are integrated and visualized to provide a comprehensive modification profile and quantitative analysis report.
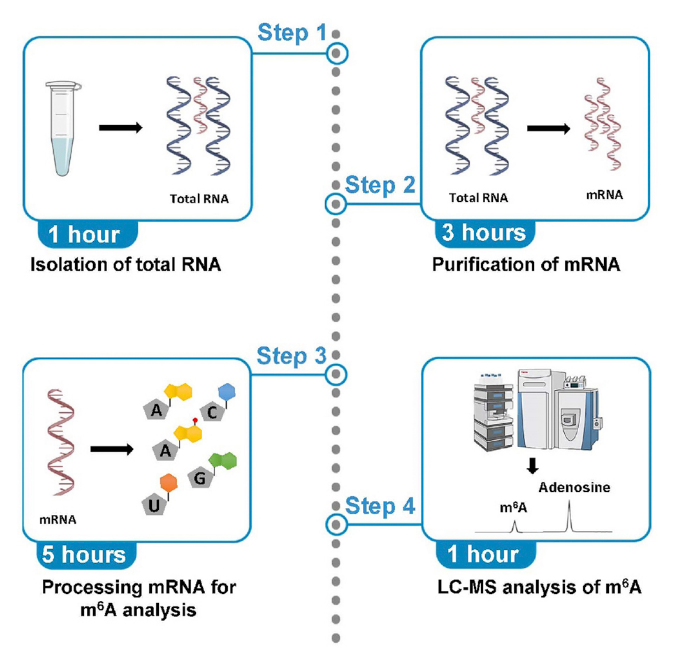
Mathur, L. et al. STAR Protocols, 2021.
Figure 2. The Workflow of m6A Modification LC-MS Analysis.
Sample Submission Suggestions
1. Sample Type
Cells, tissues, blood, and microbial samples are accepted, as well as total RNA or purified mRNA forms.
Note: Samples should be free from RNase contamination and significant degradation to ensure the accuracy of detection results.
2. Sample Storage
RNA samples should be sealed and stored in an RNase-free environment. For long-term storage, keep samples at -80°C; for short-term storage, -20°C is acceptable. Avoid repeated freeze-thaw cycles and high temperatures to prevent modification degradation.
3. Sample Transportation
Liquid samples should be transported using dry ice and cold chain conditions. Lyophilized samples can be shipped at room temperature for a short period but must be kept away from light, moisture, and high temperatures to maintain the stability of m6A modifications and overall sample integrity.
Service Advantages
1. High-Sensitivity Detection Platform
With a high-resolution LC-MS/MS system, this service enables highly sensitive detection and precise quantification of m6A modifications.
2. Comprehensive Modification Analysis Capability
It allows simultaneous analysis of m6A modification levels across different RNA sources, revealing modification distribution and dynamic variation characteristics.
3. Professional Technical Support
A team of experienced mass spectrometry and nucleic acid analysis experts provides data interpretation and methodological guidance to ensure smooth project execution.
4. Customized Research Solutions
Analytical strategies can be flexibly designed according to research objectives, supporting transcriptome integration and diverse scientific applications.
Applications
1. Epitranscriptomic Research
The m6A modification LC-MS analysis service can be used to study the distribution and regulatory features of m6A modifications at the RNA level.
2. Translational Regulation Research
By analyzing the dynamic changes of m6A modifications under different conditions, it helps reveal their regulatory patterns.
3. RNA-Binding Protein Studies
In combination with RNA-binding protein detection, this service evaluates how m6A modifications influence RBP recognition and binding affinity.
4. Species and Tissue Comparison
The m6A modification LC-MS analysis service is applied to compare the distribution characteristics and differences of m6A modifications across different species or tissues.
FAQ
Q1: Can the Analysis Distinguish between Different Methylation Types?
A1: Yes. Through high-resolution spectral analysis and standard reference comparison, multiple methylation forms such as m6A and m1A can be accurately distinguished.
Q2: Can Differences in m6A Modifications between Different Treatment Groups Be Compared?
A2: Yes. By combining TMT or Label-Free quantitative strategies, comparative analysis of m6A modification levels across multiple samples can be achieved.
Q3: Can Samples from Different Species Be Analyzed?
A3: Yes. This service is applicable to RNA samples from various sources, including human, animal, plant, and microbial samples.







