Quantitative Methyl-proteomics Service
Dynamic regulation of protein methylation influences diverse cellular pathways, from transcriptional control to metabolic regulation. Abnormal methylation is increasingly recognized as a driver of human diseases, making it an important focus in biomedical and translational research. Quantitative Methyl-proteomics Service at MtoZ Biolabs provides reliable, site-level quantification of methylation events, helping researchers uncover how this modification shapes molecular networks and biological outcomes.
Overview
Protein methylation occurs primarily on lysine and arginine residues and can exist in mono-, di-, or tri-methylated states. These subtle modifications profoundly impact chromatin remodeling, RNA processing, DNA repair, and signaling. Histone methylation has been widely studied in epigenetics, while non-histone methylation has expanded our understanding of regulatory mechanisms across the proteome.
Due to the low stoichiometry and structural diversity of methylation, conventional methods such as Western blotting or site-directed mutagenesis provide only limited resolution. Quantitative methyl-proteomics, integrating selective enrichment, LC–MS/MS, and bioinformatics, enables comprehensive mapping and dynamic profiling of methylation at the proteome scale.
Analysis Workflow
MtoZ Biolabs offers Quantitative Methyl-proteomics Service that combines enrichment of methylated peptides, advanced mass spectrometry, and computational pipelines into a unified workflow. Our process includes:
1. Protein Extraction and Digestion
Isolation of proteins from tissues, cells, fluids, or microbial samples followed by tryptic digestion.
2. Methylated Peptide Enrichment
Immunoaffinity purification using high-specificity antibodies against mono-, di-, and tri-methyl lysine and arginine, optionally coupled with HILIC or SCX fractionation for enhanced depth.
3. Quantitative LC–MS/MS Analysis
Performed on Orbitrap Fusion Lumos and Q Exactive HF platforms, with options for label-free, SILAC, or TMT/iTRAQ labeling.
4. Data Analysis and Interpretation
Site localization, quantitative comparison, motif analysis, and pathway mapping supported by state-of-the-art bioinformatics tools.
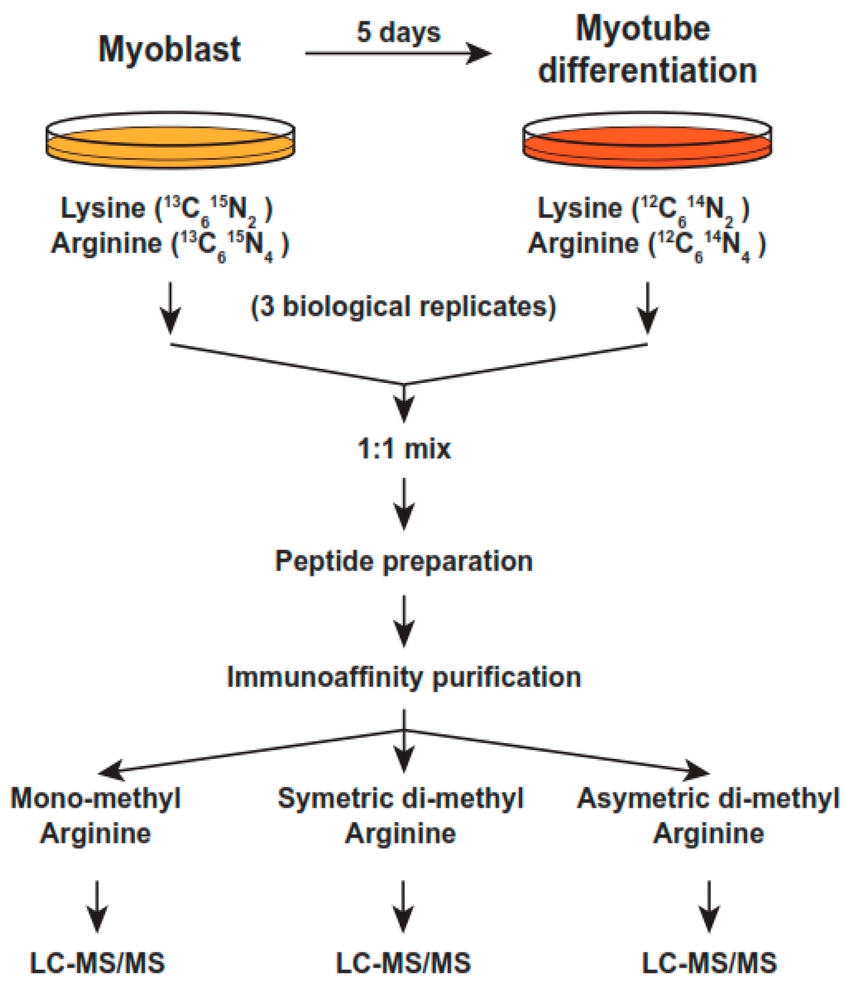
Papanikolaou, N. A. et al. Int. J. Mol. Sci. 2023.
Figure 1. SILAC-Based Workflow for Quantitative Methyl-Proteomics
Why Choose MtoZ Biolabs?
✔ Advanced Analysis Platform
MtoZ Biolabs established an advanced quantitative proteomics platform, guaranteeing reliable, fast, and highly accurate Quantitative Methyl-proteomics Service.
✔ Targeted Enrichment Strategies
Optimized antibody-based enrichment protocols maximize the recovery of methylated peptides while minimizing non-specific background.
✔ Flexible Quantitative Approaches
We offer multiple quantification options including label-free, SILAC, and TMT/iTRAQ, tailored to meet specific experimental goals.
✔ Integrated Bioinformatics Support
Our bioinformatics team delivers comprehensive data interpretation, from motif analysis to pathway mapping, enabling actionable biological insights.
✔ Reliable and Standardized Deliverables
Each project includes raw data files, processed datasets, and structured reports, ensuring transparency and supporting downstream validation or publication.
✔ One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
Sample Submission Suggestions
For optimal results in this service, we recommend the following sample amounts:
-
Serum/Plasma: 100 μL
-
Urine: 2 mL
-
Animal Tissue: 200 mg
-
Plant Tissue: 10 g
-
Microorganisms: 500 μL
-
Protein Extracts: 5 mg
-
Cells: 1 × 10⁸
Samples should be snap-frozen in liquid nitrogen and shipped on dry ice. Our technical team provides tailored guidance to ensure compatibility with downstream workflows.
Applications
Our Quantitative Methyl-proteomics Service enables researchers to move beyond simple site identification and focus on dynamic, quantitative changes in methylation under different biological states:
● Epigenetic Regulation
Quantitatively profiling histone methylation states to assess changes in chromatin accessibility and transcriptional activity across experimental conditions.
● Cancer and Neurological Disorders
Measuring differential methylation levels of regulatory proteins in tumors or neurodegenerative models to uncover disease-associated signaling alterations.
● Cellular Signaling and Stress Response
Tracking quantitative shifts in protein methylation during immune activation, cell differentiation, or environmental stress adaptation.
● Drug Mechanism and Biomarker Discovery
Comparing methylation patterns before and after drug treatment to reveal therapeutic effects, validate targets, and identify quantitative biomarkers.
● Multi-omics Integration
Incorporating quantitative methyl-proteomics data with phosphorylation, acetylation, and ubiquitylation studies to reveal coordinated PTM dynamics.
Quantitative Methyl-proteomics Service at MtoZ Biolabs delivers high-quality, site-specific datasets that illuminate the role of protein methylation in health and disease. Our integrated workflows provide actionable insights for epigenetics, signaling, and translational research.
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) services provider, also offers comprehensive post-translational modification (PTM) proteomics solutions, including phosphorylation, acetylation, ubiquitination, SUMOylation, and more. With cutting-edge platforms and proven expertise, we help researchers uncover regulatory mechanisms and accelerate biomedical innovation. Contact us today to explore how our PTM services can advance your work.







