Novel Post-translational Modifications Analysis Service
Proteins are the functional workhorses of all biological systems, and their activities are intricately modulated through a diverse range of post-translational modifications (PTMs). While canonical PTMs such as phosphorylation, acetylation, and ubiquitination have been extensively studied, an expanding array of novel post-translational modifications has emerged, revealing new regulatory mechanisms in cellular signaling, metabolism, stress responses, and disease development. These unconventional modifications, including acylation, lipidation, redox-based modifications, and amino acid–derived chemical alterations, represent frontier areas of proteomics research, yet their detection and characterization remain technically challenging due to low abundance, transient nature, and chemical diversity.
Services at MtoZ Biolabs
MtoZ Biolabs provides a comprehensive Novel Post-translational Modifications Analysis Service to enable precise identification and quantification of rare or unconventional PTMs using advanced mass spectrometry–based proteomics. By integrating optimized sample enrichment protocols, high-resolution LC-MS/MS platforms, and specialized bioinformatics workflows, we help researchers decode complex PTM landscapes and uncover new layers of biological regulation.
1. Novel Post-translational Modifications Analysis of Target Protein
For protein-specific investigations, we offer targeted characterization of novel PTMs on individual proteins. This approach allows accurate identification of modification sites and quantitative assessment of modification states under defined biological conditions.
2. Novel Post-translational Modifications Proteomics Analysis
For broader discovery-driven studies, we perform proteome-wide analysis to systematically profile unconventional PTMs across complex samples. High-resolution mass spectrometry combined with bioinformatics mapping reveals global modification patterns, pathway involvement, and regulatory networks.
To comprehensively support the exploration of emerging PTMs, MtoZ Biolabs has established a specialized analytical framework covering multiple modification categories:
💠Acylation & Lipidation Analysis Service: Investigating diverse acyl and lipid conjugations such as succinylation, malonylation, crotonylation, and palmitoylation that regulate protein stability and membrane association.
💠Redox-related Modifications Analysis Service: Profiling cysteine-centered and oxidative modifications including S-nitrosylation, S-sulfenylation, S-glutathionylation, and disulfide bond dynamics that shape redox signaling.
💠Glycosylation and Related Modifications Analysis Service: Characterizing N-, O-, and C-linked glycosylation, glypiation, and glycan-related variants essential for protein folding, recognition, and immune regulation.
💠Methylation and Related Modifications Analysis Service: Quantifying mono-, di-, and tri-methylation of lysine and arginine residues and exploring crosstalk with acetylation and phosphorylation pathways.
💠Amino Acid-derived Modifications Analysis Service: Detecting metabolite-driven PTMs such as lactylation, succinylation, β-hydroxybutyrylation, and citrullination that connect metabolism with epigenetic regulation.
💠Ubiquitin-like Modifications Analysis Service: Examining conjugation systems including SUMOylation, NEDDylation, and Pupylation that modulate protein turnover and signal transduction.
Analysis Workflow
1. Sample Preparation: Protein extraction, quantification, reduction, alkylation, and enzymatic digestion are performed under optimized conditions to preserve novel PTMs.
2. Modified Peptide Enrichment: Chemical derivatization, affinity capture, or ion exchange techniques are applied to selectively isolate modified peptides from complex mixtures.
3. LC-MS/MS Analysis: High-resolution mass spectrometric detection is used for accurate identification and quantification of modified peptides.
4. Data Processing and Interpretation: Database searching with variable modification parameters, spectral validation, and precise site mapping are conducted to ensure data reliability.
5. Bioinformatics Integration: Pathway enrichment, motif discovery, and functional analysis are performed to connect PTM profiles with biological mechanisms.
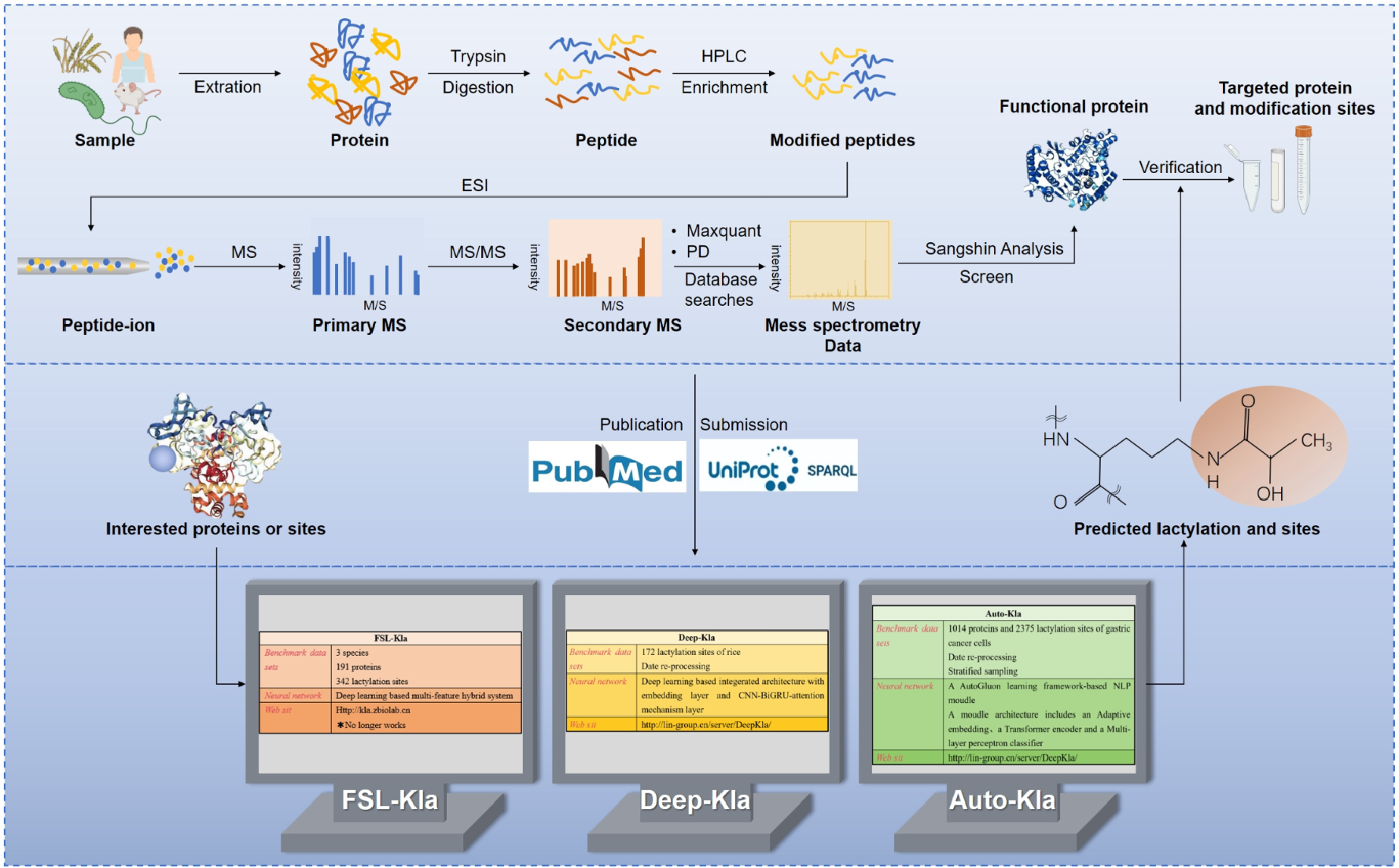
Liu, J. et al. Biomolecules. 2024.
Figure 1. General Workflow for the Detection of Novel Post-translational Modifications (Example: Lactylation)
Sample Submission Suggestions
1. Sample Type: Cell lysates, tissue homogenates, serum/plasma, or purified proteins.
2. Amount Required: Minimum 500 µg total protein per sample.
3. Storage and Transport: Samples should be snap-frozen in liquid nitrogen and shipped on dry ice.
*Note: For unusual sample types or low-yield materials, contact MtoZ Biolabs for tailored pre-processing guidance.
Service Advantages
✅
✅High Data Quality: AI-assisted data analysis ensures accurate identification, quantification, and reproducibility.
✅Comprehensive Workflow: End-to-end service from sample processing to bioinformatics interpretation.
✅Professional Expertise: A multidisciplinary team of proteomics and PTM specialists provides technical consultation and data interpretation.
✅Customizable Solutions: Flexible experimental designs tailored to different sample types, species, and research objectives.
Applications
1. Epigenetic and Transcriptional Regulation
Study the role of emerging acylations and other PTMs in chromatin remodeling, histone modification, and gene activation.
2. Metabolic Pathway Mapping
Correlate novel PTMs such as succinylation and lactylation with metabolic flux and energy regulation.
3. Disease Mechanism Studies
Explore PTM dysregulation in cancer, cardiovascular disease, diabetes, and neurodegenerative disorders.
4. Drug Discovery and Therapeutic Targeting
Identify PTM-associated enzymes and pathways that serve as potential drug targets.
5. Biomarker Discovery
Detect novel PTM patterns as diagnostic or prognostic markers in precision medicine.
FAQ
Q1: Can you discover entirely unknown PTMs (never reported before)?
Yes. Our open-search strategy and manual validation allow the detection of unexpected mass shifts. However, biological validation (e.g. mutagenesis or orthogonal assays) is often recommended.
Q2: Can multiple PTM classes be profiled in the same experiment?
Yes, if enrichment strategies are compatible. In many cases, we perform "split-enrichment" parallel workflows followed by integrated data annotation.
Q3: What about false positives for rare modifications?
We employ strict FDR control, diagnostic ion filtering, manual spectral check, and replicate consistency to reduce artifacts.







