S-Nitrosylation Site Mapping Service
Based on a high-resolution mass spectrometry (LC-MS/MS) platform and highly specific chemical labeling technology, MtoZ Biolabs has launched the S-nitrosylation site mapping service which enables systematic identification and quantitative analysis of S-nitrosylation modification sites in proteins. This service integrates Biotin-switch, TMT labeling, and enrichment strategies to precisely capture S-NO-modified peptides and obtain high-confidence modification site information through mass spectrometry detection. The final report provides data on the distribution, abundance changes, and functional annotation of protein S-nitrosylation modifications, offering reliable data support for redox regulation and protein function studies.
What Is S-Nitrosylation?
Protein S-nitrosylation is a reversible cysteine redox modification that involves the covalent attachment of a nitric oxide (NO) molecule to the thiol group (-SH) of cysteine residues, forming an S-NO bond. This modification is widely present in animal, plant, and microbial systems, regulating protein conformation, activity, and interactions, and participating in various biological processes such as cell signaling, metabolic regulation, and stress response. With the advancement of mass spectrometry technology, site-specific identification and quantitative analysis of S-nitrosylation have become essential tools for studying NO signaling mechanisms and redox regulation, with broad applications in biochemistry, molecular biology, pharmacology, and environmental toxicology.
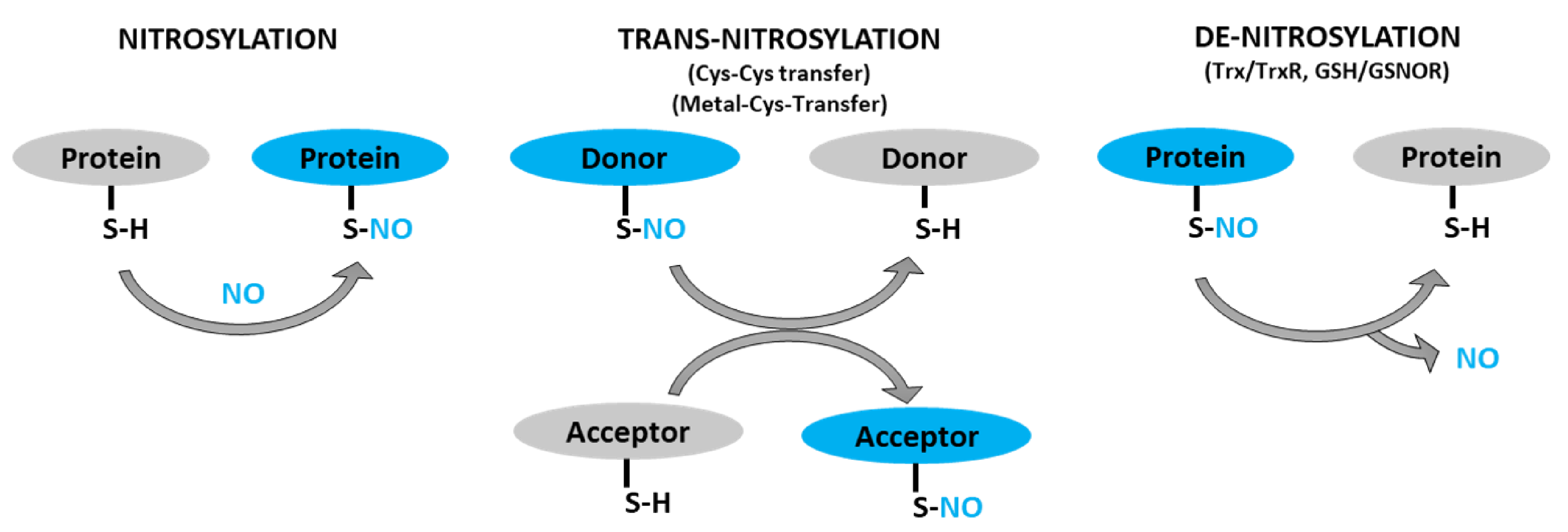
Sharma, V. et al. International Journal of Molecular Science, 2021.
Figure 1. Schematic Representation of Protein S-Nitrosylation, Transnitrosylation, and Denitrosylation.
Services at MtoZ Biolabs
1. Target Proteins S-Nitrosylation Site Identification
MtoZ Biolabs can accurately identify S-nitrosylation sites on target proteins, analyzing their modification types and locations. Through high-resolution LC-MS/MS platforms, we can compare site variations under different conditions, revealing the impact of nitrosylation on protein function.
2. Proteomics S-Nitrosylation Site Identification
By combining enrichment strategies with large-scale mass spectrometry, MtoZ Biolabs can identify and quantify multiple S-nitrosylation sites at the proteomic level. This analysis helps construct a comprehensive S-nitrosylation modification map, supporting regulatory mechanism research and biomarker discovery.
Analysis Workflow
1. Protein Extraction and Sample Preparation
Total proteins are extracted from tissue, cell, or biofluid samples under antioxidant conditions to preserve the native S-NO modification state.
2. Chemical Labeling and Modification Conversion
The Biotin-switch or TMT labeling strategy is used to convert S-NO modifications into detectable forms.
3. Enrichment and Purification
Affinity capture or chemical enrichment methods are applied to selectively isolate S-nitrosylated peptides.
4. Mass Spectrometry Detection and Data Acquisition
High-resolution LC-MS/MS is employed to detect enriched peptides and acquire fragment spectra for site identification.
5. Data Analysis and Reporting
Database searching and algorithm-based analysis are performed to generate comprehensive reports including site identification, quantitative results, and functional annotation.
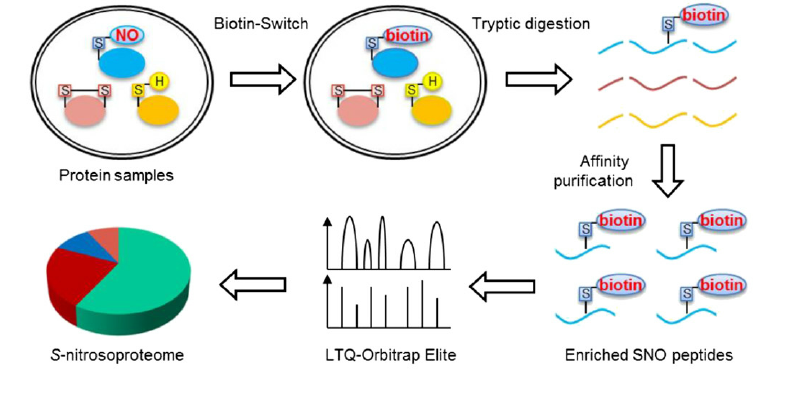
Xu, J L. et al. Plant Physiology, 2015.
Figure 2. Site-Specific Identification of Endogenously S-Nitrosylated Proteins.
Sample Submission Suggestions
1. Sample Type
Cell, tissue, serum, plasma, and microbial samples are acceptable, and protein extracts or lyophilized powders are also supported.
Note: Avoid the use of strong oxidants or reductants during sample preparation to preserve the native S-NO modification state.
2. Sample Storage
Samples should be sealed, protected from light, and stored at -80°C for long-term preservation or at -20°C for short-term storage. Avoid repeated freeze-thaw cycles and prolonged air exposure to prevent degradation of S-nitrosylation modifications.
3. Sample Transportation
Liquid samples should be transported under cold-chain conditions. Lyophilized samples can be shipped at room temperature for short periods but should be protected from heat, humidity, and strong light to ensure modification stability and analytical accuracy.
Service Advantages
1. High-Sensitivity Mass Spectrometry Platform
Utilizing a high-resolution LC-MS/MS system, precise detection and quantification of S-nitrosylation modifications are achieved.
2. High-Specificity Labeling and Enrichment
Biotin-switch or TMT strategies are applied to significantly enhance the capture efficiency of modified peptides.
3. Comprehensive Site Characterization
Systematic identification of S-nitrosylation site distribution and abundance changes supports both qualitative and quantitative analyses.
4. Customized Research Design
Experimental workflows can be flexibly tailored to project requirements, supporting integrated studies with other post-translational modifications.
Applications
1. Signal Transduction Research
The S-nitrosylation site mapping service can be used to investigate the regulatory role of S-nitrosylation in cellular signaling pathways.
2. Metabolic Regulation Analysis
By detecting modifications on metabolic enzymes, this service helps assess their impact on energy metabolism and material conversion.
3. Redox Homeostasis Studies
The S-nitrosylation site mapping service enables the exploration of dynamic S-nitrosylation changes under different oxidative conditions.
4. Protein Function Research
Through precise site identification, researchers can examine how S-nitrosylation affects protein structure and enzymatic activity.
FAQ
Q1: How Can Sample Modification Loss Be Prevented during Preparation?
A1: It is recommended to perform all procedures under low-temperature, light-protected, and antioxidative conditions. Avoid using reducing agents such as DTT or β-ME to maintain the stability of the modification state.
Q2: Can both Modification Sites and the Affected Protein Functions Be Identified?
A2: Yes. Based on site identification, we integrate functional annotation and pathway enrichment analyses to reveal the potential regulatory roles of S-nitrosylation in protein function.
Q3: Is the mass Spectrometry Sensitivity Sufficient to Detect Low-Abundance Modifications?
A3: With a high-resolution LC-MS/MS system and high-specificity enrichment strategy, even low-abundance S-nitrosylated peptides can be reliably detected.







